-
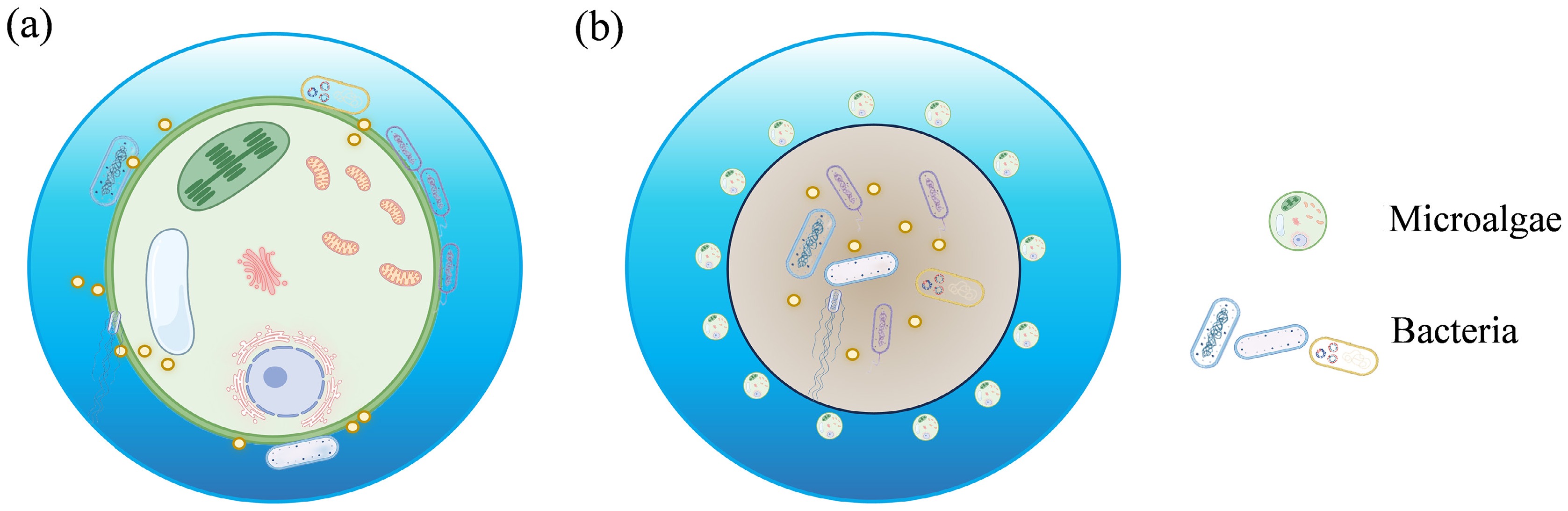
Figure 1.
Schematic diagram of the spatial structure of the (a) phycosphere, and (b) MBGS.
-
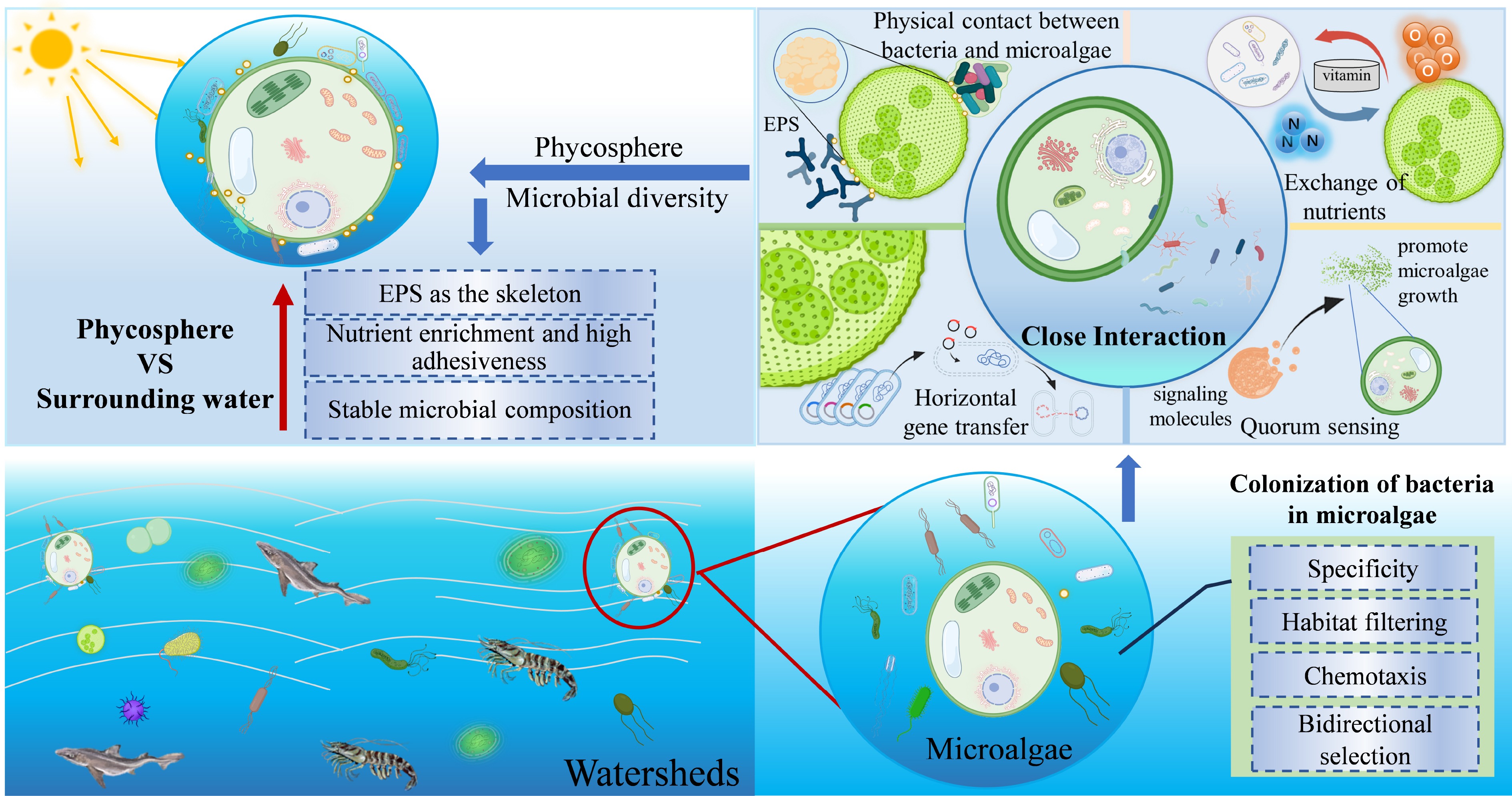
Figure 2.
Bacterial colonization and characteristics of the phycosphere environment.
-
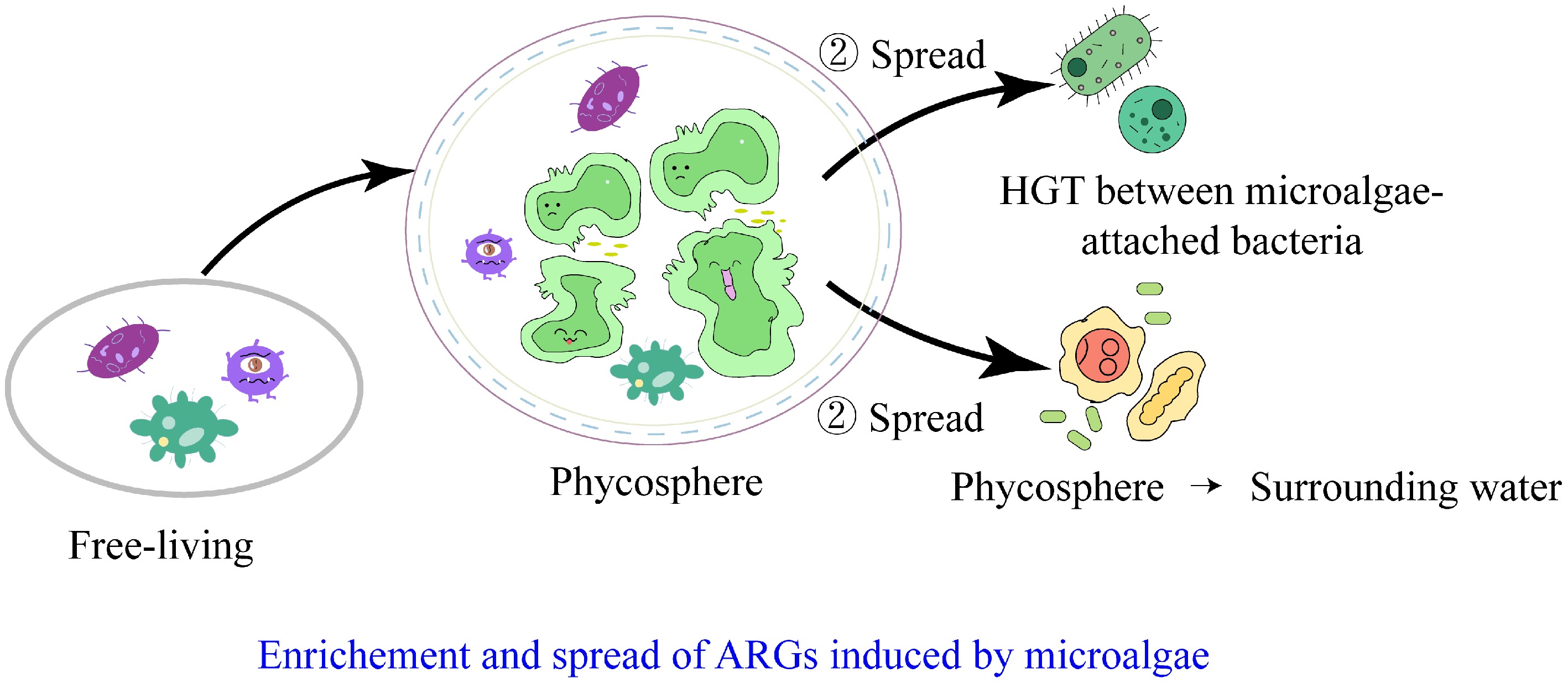
Figure 3.
Pathways of ARGs enrichment and spread induced by microalgae in watersheds.
-
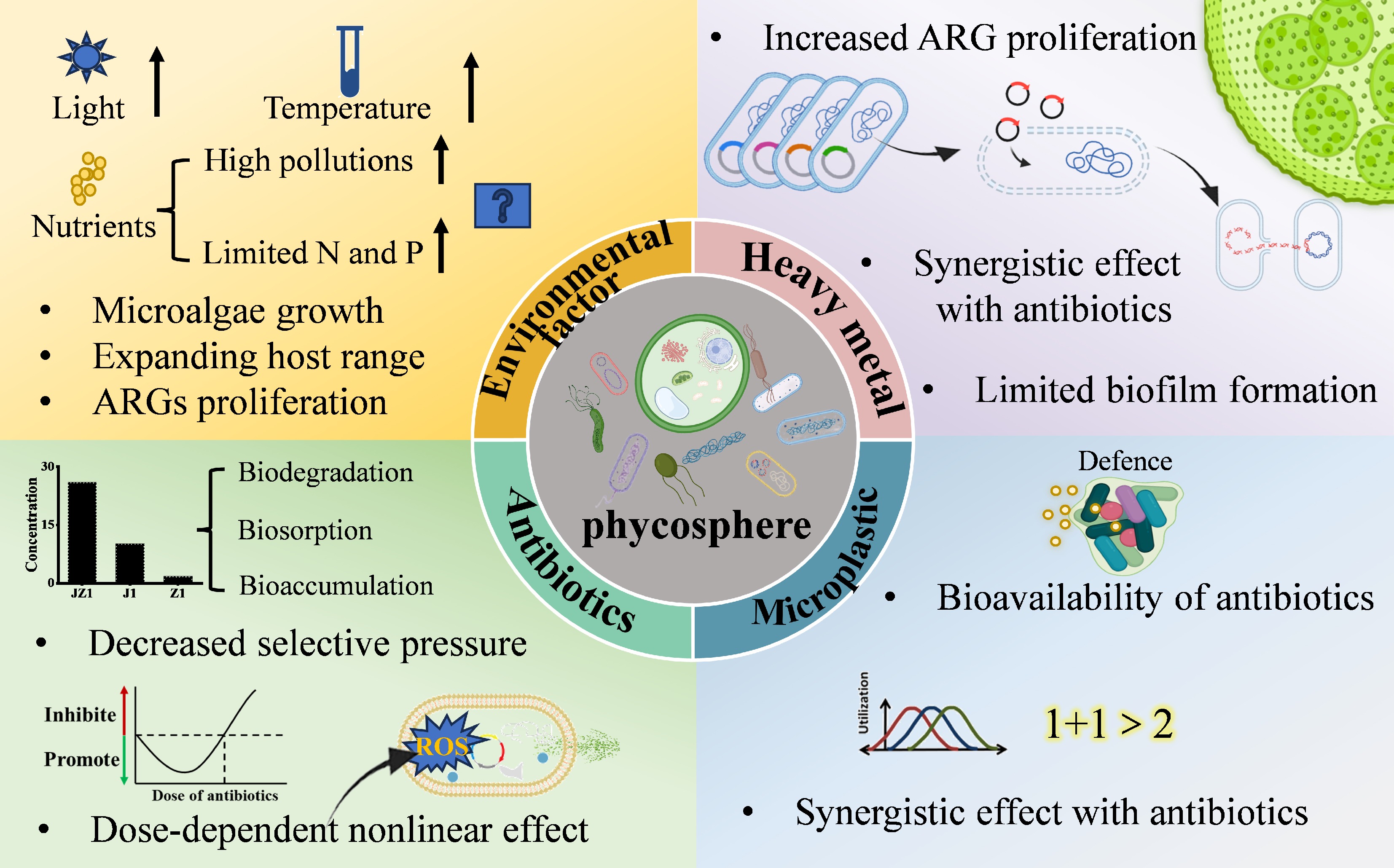
Figure 4.
The effects of contaminants on antibiotic resistance genes in the phycosphere.
-
Order Microalgae species ARGs types Exogenous pollutants Sampling point/bacterial source Article type Topic Ref. 1 Cyanobacteria intI1, 16S rRNA, sulfonamide resistance genes (sul1, sul2, sul3, sulA), tetracycline resistance genes (tetA, tetB, tetD, tetE, tetJ, tetK, tetM, tetO, tetQ, tetS, tetW), macrolide resistance genes (ermB, ermC, ereA, ereB, mphA, mphB), quinolone resistance genes (qnrB, qnrS, aac(6′)-Ib), β-lactam resistance genes (blaTEM, blaOXA, blaOXY, blaSHV, blaCTX-M), and aminoglycoside resistance genes (strA, strB) Cyanobacteria bloom Taihu Lake, China Sampling from
surface waterAntimicrobial resistance in the phycopshere [52] 2 / Sulfonamide resistance genes (sul1, sul2, sul3), tetracycline resistance genes (tetA, tetB, tetC, tetG, tetM, tetO, tetQ, tetW, tetZ), quinolone resistance genes (gyrA, qnrA, qnrB, qnrS), macrolide resistance genes (ermB, ermC, ermF), chloramphenicol resistance genes (cat1, floR, cmlA), β-lactams resistance genes (blaNDM1, blaIMP4, blaCTX-M, blaSHV), integron integrase genes (intI1, intI2), transposases (tnpA-01, tnpA-02, tnpA-03, tnpA-04, tnpA-05, tnpA-07, IS613), 16S r RNA River contamination Ba River, China Sampling from
surface waterAntimicrobial resistance in the phycopshere [51] 3 Cyanobacteria 296 primer pairs that targeting one 16S gene, 285 major ARGs and 10 MGEs (1 clinical class 1 integron, 1 class 1 integron and eight transposases) Cyanobacterial bloom Taihu Lake, China Sampling from
surface waterAntimicrobial resistance in the phycopshere [36] 4 Green tide Metagenomic sequencing Green tide bloom Yellow Sea, China Sampling from
surface waterAntimicrobial resistance in the phycopshere [37] 5 / / / Lake Mochou,
ChinaSampling from
surface waterBacterial colonization and assembly in microalgae [63] 6 / / / North Sea island Helgoland Sampling from
surface waterBacterial colonization and assembly in microalgae [23] 7 Ulva sp., Saccharina sp., Grateloupia sp., and Gelidium sp. / / Weihai, China Sampling from
surface waterBacterial colonization and assembly in microalgae [54] 8 Chlorella pyrenoidosa Two sulfonamide resistance genes (sul1 and sul2), 15 tetracycline resistance genes (tet(36)-01, tet(36)-02, tetA-02, tetG-01, tetG-02, tetL-02, tetM-01, tetM-02, tetQ, tetR-01, tetR-02, tetS, tetT, tetW-01 and tetX), and five MEGs (intI1, tnpA-03, tnpA-04, tnpA-05, tnpA-07) Silver nanoparticles (AgNPs)
Hematite nanoparticles (HemNPs)
Tetracycline
SulfadiazineChicken farm wastewater Laboratory experiment Antimicrobial resistance in the phycopshere [75] 9 Chlorella pyrenoidosa Sulfonamide resistance genes (sul2, sul1, dfrA1), tetracycline resistance genes (tetX, tetG-02, tetG-01, tetM-01, tetM-02, tetA-01, tet(36)-02, tetL-02, tetA-02), aminoglycoside resistance gene (aadA2–03, aadA1, aadA2–01, aadA2–02, aadA-02, aadA-01, strB, aadA5–01, aac(6′)-Ib-3, aadA5–02, aac(6')-Ib-1, aac(6')-Ib-2, aadE, aac (6')-II, aphA1, aadA9–02), MGEs (tnpA-04, intI1, tnpA-03, tnpA-07) Sulfadiazine Soil leachate near poultry farms Laboratory experiment Antimicrobial resistance in the phycopshere [70] 10 Chlorella vulgaris sul1, sul2, intI1 Sulfadiazine Wastewater from fish ponds Laboratory experiment Antimicrobial resistance in the phycopshere [71] 11 Chlorella pyrenoidosa Sulfonamides resistance genes (sul1 and sul2), phenicol resistance genes (floR, fexA, fexB, optrA, cfr, and pexA), integrase and transposase (intI1, IS613, tnpA01, tnpA02, and tnpA03) Polylactic acid MP
FlorfenicolDonghu lake and fish ponds Laboratory experiment Antimicrobial resistance in the phycopshere [72] 12 Auxenochlorella pyrenoidosa Metagenomic sequencing Contents of N and P Donghu lake Laboratory experiment Antimicrobial resistance in the phycopshere [61] 13 Chlorella vulgaris tetQ, tetE, sul1, sul2, strA, strB, blaOXA, blaPSE / Effluent from the sewage treatment plant Laboratory experiment Antimicrobial resistance in the phycopshere [10] 14 Chlorella vulgaris sul1, sul2, sul3, intI1, 16S rRNA gene Sulfamethoxazole Jinggong Lake Laboratory experiment Antimicrobial resistance in the phycopshere [16] 15 Chlorella vulgaris, Scenedesmus acuminatus, Chlamydomonasnoctigama, Chlorolobion braunii, Raphidocelis subcapitata Tetracycline resistance genes (tetA, tetB, tetC, tetZ), quinolone resistance genes (gyrA, qnrA, qnrB, qnrS), macrolide resistance genes (ermB, ermC, ermF), chloramphenicol resistance genes (catA1, floR, cmlA), aminoglycoside resistance genes (aadA, aacC, aph(2’)-Id), and β-lactam resistance genes (blaCTX-M, blaSHV, blaTEM), intI1, intI2, 16S rRNA gene Rivers with varying
degrees of pollutionQishui River Laboratory experiment Antimicrobial resistance in the phycopshere [17] 16 Chlorella vulgaris 16S rRNA, tetracycline resistance genes (tetA, tetB, tetC, tetG, tetM, tetO, tetQ, tetW, tetZ), quinolone resistance genes (aaC(6')-Ib, gyrA, oqxAB, qepA, qnrA, qnrB, qnrC, qnrD, qnrS), aminoglycoside resistance genes (aph(2')-Id), β-lactam resistance genes (blaSHV), macrolide resistance genes (ermC), chloramphenicol resistance genes (floR), sulfonamide resistance genes (sul2), mobile genetic elements (intI1, intI2, IS613, tnpA-02, tnpA-03) Tetracycline
CiprofloxacinWei River Laboratory experiment Antimicrobial resistance in the phycopshere [50] 17 Chlorella pyrenoidosa pEASY-T1 plasmid Contents of N / Laboratory experiment Antimicrobial resistance in the phycopshere [35] 18 Cyanobacteria Metagenomic sequencing Temperature and phosphorus concentration The water and sediments of the Three Gorges Reservoir Laboratory experiment Bacterial colonization and assembly in microalgae [84] 19 / / / / Laboratory experiment Bacterial colonization and assembly in microalgae [15] 20 Diatom / / / Laboratory experiment Bacterial colonization and assembly in microalgae [21] Table 1.
The summary of the microalgae and ARGs related to the phycosphere in the previous studies
Figures
(4)
Tables
(1)