-
Plastic is a unique material that facilitates all aspects of our lives. Due to its low density, durability, malleability and high resistance to corrosion, its use is rapidly increasing[1]. Moreover, it is available at a low cost to consumers and therefore is replacing other materials such as wood, metal, and glass[2]. To meet the increasing demand, millions of tons of plastic are produced every year. According to a 2017 report, global plastic production has reached 359 million metric tons[3]. If the current trends of production and use goes unchecked, without proper management strategies, terrestrial and aquatic ecosystems may end up overloaded with plastic. Since plastic is a recyclable material[4], only 9% is reclaimed, 12% is incinerated and 79% ends up in water and soil bodies after being dumped in landfill[5]. The plastic debris arriving in these environments has now begun to cause serious problems in ecosystem functions, biodiversity and food web exchanges. The environmental pollution caused by the production and the disposal of petrochemical derived plastics has raised increasing concern[1,6,7]. Therefore, profound waste management strategies are required that can minimize plastic accumulation in the environment.
Different approaches are being utilized to handle the increasing scourge of plastic pollution in the environment. Recycling of plastic is an approach to recover plastic and reduce plastic pollution. However, recycling is neither not always possible nor economically feasible for all types of plastics[8]. Different kinds of plastics include different chemical additives and colorants that cannot be recycled together. Mass burning technology is another widely used method to decrease plastic mass. However, due to serious environmental consequences[9], its adoption cannot be recommended. Burning produces smoke which includes acid gases, carcinogenic dioxin, particulates, heavy metals, and nitrogen oxide. These gases are poisonous to the environment. Despite the adoption of various disposal methods, the environmental persistence of plastic waste and their potential for pollution have not yet been solved[10].
Plastic is a long polymeric material that is formed by the condensation of small monomers in a repeated manner. Broadly, plastic is categorized into two types, petrochemical-plastic and bio-plastic. Petrochemical-plastic is synthesized from hydrocarbons during complex chemical reactions, and generally includes highly persistent forms such as polyethylene, polystyrene, nylons, polyurethanes, and polyesters. However, bio-plastic is produced from natural renewable and biodegradable raw materials such as sugars, starch, and cellulose obtained from plants and other agricultural sources. The commonly used bio-plastics are poly-3-hydroxybutyrate, polyhydroxyvalerate, polylactic acid, and poly ε-caprolactone[11,12]. Given the high cost and limited performance[13−16], bio-plastic use does not seem a feasible solution to plastic pollution. Moreover, a complete ban or restriction on the use of petrochemical-plastic is expected to negatively impact the economy of a country[5].
Compared with all these chemical or physical methods, biodegradation could offer a safe and eco-friendly plastic waste management solution[17]. Biodegradation is the process by which microorganisms, including algae, fungi and bacteria, are involved in the degradation of polymers[18]. Despite the fact that biodegradation is the most suitable method for plastic waste management, large-scale application of this process is still in its infancy.
In this review, we intend to draw the attention of the scientific community towards the design of a more efficient biological method for the degradation of petrochemical plastics. The identification of plastic degrading microbes and their enzymes, together with advancement in fundamental biology and other related disciplines offer promising starting points for the development of methods to produce bio-catalysts for the biodegradation of plastics. Here, we give an overview of the major applications and advancements of biocatalyst technology. We also discuss different stages for the development of bio-catalytic plastic degrading technology and the major issues related to each stage. The establishment of a standardized method of bio-catalyzed plastic degradation would help to address the looming environmental threat posed by plastic accumulation.
-
Several microbes and their corresponding enzymes that are capable of plastic degradation have been identified[19−28]. The flow chart of the various steps involved in the process of bio-catalyzed plastic degradation technology are depicted in Fig. 1. In general, three steps are involved in the bio-catalyzed degradation of plastics in soils[29]: (1) microbial colonization on the surface of the plastic, (2) enzymatic depolymerization of the plastic to low molecular weight fragments by microbes, and (3) microbial utilization of the low molecular weight fragments leading to the ultimate degradation of the polymer. Both extracellular and endocellular microbial enzymes participate in the degradation process. The extracellular enzymes depolymerize the plastic into its monomers, which are then utilized by the microbial cells and mineralized to end-products such as CO2, H2O and biomass[30]. The bio-catalyzed degradation of plastics proceeds actively under natural environmental conditions. It is frequently limited by tight regulation of enzymes that often reduces plastic degradation efficiency under most environmental conditions[27,28]. The degradation process is also controlled by many factors such as polymer characteristics, type of organism, and reaction conditions[31]. Normally, high molecular weights result in a sharp decrease in solubility, making them unfavorable for microbial attack and its utilization[31]. Many scientists suggested that for efficient biodegradation, a certain type of pretreatment is needed to break down the complex structure of the plastic[31−33]. Furthermore, the surface conditions (e.g. surface area, hydrophilic, and hydrophobic properties), the first order structures (e.g. chemical structure, molecular weight and molecular weight distribution) and the higher order structures (e.g. glass transition temperature, melting temperature, modulus of elasticity, crystallinity and crystal structure) of polymers are crucial factors influencing the biodegradation process[32].
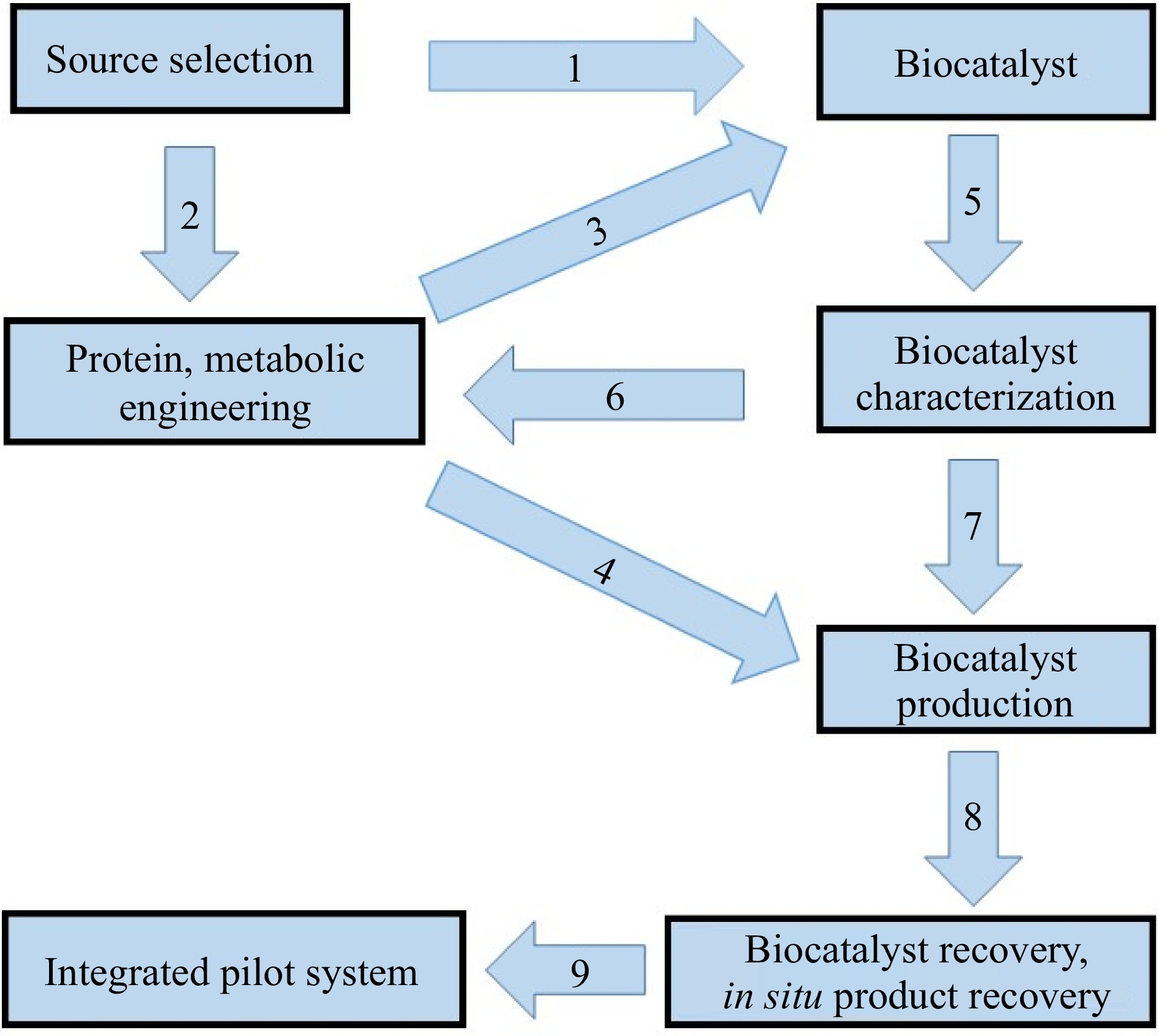
Figure 1.
Diagram of the steps involved in biocatalyzed plastic degradation. The source is a microorganism (bacterium or fungus) in the proper amount with stable enzymatic and growth activities and low cost. Biocatalyst(s) would be isolated from the source (1) and if needed this source can be improved through genetic engineering technology (2) to be able to secrete only a specific enzyme that could be used as a biocatalyst (3). In the case of whole cell catalysis, the source can be used directly as a biocatalyst. Recombinant DNA technology could be employed to generate mutant strains with an increased production of desired enzymes and a minimum production of undesirable enzymes (4). The biocatalyst should be characterized (5) and in some cases modified to improve the catalytic efficiency (6), so that it can be produced in large amounts (7). The recovery of the biocatalyst (8) should be ensured in a well-developed and sophisticated bioreactor (9).
-
Polyethylene (PE) is one of the most common types of plastic. The biodegradation of PE by both bacterial and fungal enzymes have been identified. These enzymes include laccases, manganese peroxidase and lignin peroxidases[33−37]. However, the biocatalysts could not achieve efficient biodegradation due to their high redox potential requirements[33,37]. The lack of hydrolysable functional groups in the PE backbone hampers the biodegradation process[32,37].
Polyurethane
-
Polyurethane (PU) is a heteropolymer formed by the condensation of di- or poly-isocyanate and polyols linked by urethane linkages[38]. Depending on the nature of the polyols used for the polycondensation reaction, two different types of PUs i.e. polyether and polyester can be prepared. Several studies have identified that microbial enzymes of bacterial and fungal domains are capable of depolymerizing PU[24,25,28,31,39−47]. Ureases, esterases and proteases are the some of the important enzymes that can hydrolyze the urethane and ester bonds in the PU[28,31,46]. While previous studies have observed extensive surface degradation and tensile strength loss in PU films by microbes[39−43], the weight loss observed was too low, indicating that the catalytic efficiencies of enzymes were not high[24,47].
Polystyrene
-
A very efficient protocol for the biodegradation of polystyrene (PS) was developed by Nakamiya et al.[19]. They used a purified hydroquinone peroxidase isolated from Azotobacter beijerinckii HM121 in a two-phase system consisting of dichloromethane and water. PS was reported to be converted into water soluble daughter products within 5 min of reaction at 30 °C. However, this protocol has not been implemented for recycling of PS.
Polyethylene terephthalate
-
Polyethylene terephthalate (PET) polymer is synthesized from terephthalic acid and ethylene glycol by an esterification reaction[48]. PET is widely used in the textile industry and packaging materials. This polymer has different degrees of crystallinity which strongly affects its biodegradability[49]. A number of enzymes capable of biodegrading PET have been identified[50−53]. Recently, polyethylene terephthalate-degrading enzyme (PETase) has been isolated from the bacterium that exhibits both lipases and cutinase activity[27]. The biodegradation of amorphous PET to its monomers in an enzyme reactor has been demonstrated by Barth et al.[54]. Enzyme engineering at the catalytic site was found to improve the PET degrading efficiency by 20%[55,56]. Studies suggest that this enzyme can be utilized for a biotechnical recycling process of PET waste. The majority of these plastics can be degraded to a certain extent if appropriate enzymes are applied at an optimum concentration within a specific environment[17]. However, to date there are no reports on the industrialized scale application of PET degradation.
-
Biocatalysts are natural catalysts, such as enzymes or microbial cells that can speed up biochemical processes without being consumed. Biocatalyst technology is an interdisciplinary field using free-enzymes or whole-cell biocatalysts for large-scale bioprocesses[57]. Biocatalyst technology is an important component of sustainable industrial development[58]. Its applications range from in-house applications to food technology and agriculture, textile industry, genetic engineering, high-throughput screening, pharmaceutical industry and other emerging technologies[58,59]. Biocatalyst technology has been recognized to improve the economics of existing processes by reducing operating costs up to 90% by reducing energy consumption and use of raw material resources[60]. In polymer sciences, an important application of biocatalyst technology is the use of microbial isolated enzymes in vitro for polymer synthesis[61−63]. The success of biocatalysis in polymer synthesis has been attributed to several characteristics of enzymes. These include their high recyclability rate, high substrate conversion efficiency and their highly selective nature[61]. A number of studies have reported the synthesis of polyesters using bio-catalysts[62−64]. On the other hand, efficiency of biocatalyzed polymer degradation has been studied for biodegradable polymers[65,66]. For petrochemical-based plastics, biocatalysts have been identified from different microbial sources[26-28,31-33]; however, it has not been practically adopted. In conclusion, biocatalyst technology can serve as a strategy to help remediate accumulated plastic across ecosystems.
-
There are various steps that are involved in developing biocatalyzed plastic degradation technology. Here we discuss these steps in detail.
Source selection
-
The critical step in biocatalyzed plastic degradation is the screening and selection of the source for the biocatalyst. In many cases, the source is a microorganism such as a bacterium or fungus. The desired qualities for a microorganism to be selected as a source for biocatalyzed plastic degradation technology includes a high growth rate on a low-cost medium, with stable physiological characteristics and high enzyme activity[58]. The availability of the genetic information of the source is also very important for its utilization in biocatalyzed plastic degradation technology[59]. Genetic information helps in the manipulation of genetic material to improve the efficiency of these processes. Recombinant DNA technology could be employed to generate mutant strains with an increased production of desired enzymes and a minimum production of undesirable enzymes[67]. Metagenomic approaches could be employed for the identification of novel proteins or enzymes in microorganisms, which cannot be grown under laboratory conditions[68]. Some studies suggest that thermophilic microbes are beneficial as a source because of reduced chances of inactivation of the enzymes. Their adaptability to survive at high temperatures enhances their lifetime during subsequent utilization[69]. Substrate specificity of the microbe is also an important characteristic[31]. Microbes with a broad range of substrate degrading activity are a preferred choice[70]. Novel sources of potentially useful microbes include Roseateles depolymerans, an aerobic and mesophilic bacterium that can degrade a wide range of polymers such as polybutylene succinate (PBS), polycaprolactone (PCL)[70], polybutylene carbonate (PBC), poly(butylene succinate)-co-(butylene adipate) (PBSA). Thermobifida alba is a thermophilic bacterium that produces a thermostable esterase enzyme[71]. Pseudozyma antarctica is an extremophile originally isolated from Antarctica that had the widest range of substrate specificity and could effectively biodegrade PCL, PLA, PBS, and PBSA[72]. Aspergillus tubingensis, a filamentous fungus isolated from a rubbish dump in Pakistan is highly efficient at degrading polyurethane polymers[28].
Biocatalyst isolation, purification and characterization
-
After the fermentation process is completed, subsequent recovery of biocatalysts usually follows a number of different paths commonly employed for protein isolation. The isolation process is very sensitive with the risk that the enzyme could become unstable if not handled properly. Information about whether enzyme localization is intracellular or extracellular, is important for selecting and designing an isolation or recovery method for extracting the biocatalyst from the medium[27,46]. Once the pure biocatalyst has been obtained, the next task is to fully characterize the biocatalyst for its chemo-, regio-, and stereo-selectivity, kinetic properties, substrate specificity, thermodynamics and optimum conditions of reaction[17]. Exploring the structure of the biocatalyst is an important task for understanding its mechanism of action. Structural techniques including X-ray diffraction, neutron diffraction, and electron microscopy are able to determine the molecular structure of the biocatalysts[55,56]. The rate of enzyme catalyzed reaction is dependent on the operating environmental conditions. Therefore, experimental optimization may produce a better understanding of the enhanced function of the biocatalyst for industrial application[57]. Biochemical and biophysical studies will be useful to provide additional information about the properties and activity of the biocatalysts.
Biocatalyst engineering
-
Biocatalysts obtained from living systems have evolved towards their natural roles and functions; thus, in their native state they are not suitable for industrial biotechnology. For industrial biotechnology, enzymes should be more active, specific and selective towards their substrates[58]. To meet the requirements of industrial biotechnology, in most cases these biocatalysts must be bioengineered[73]. Biocatalyst or enzyme engineering involves the optimization of enzymes by modification to the structure and/or properties of the enzyme. Biocatalyst engineering involves two general approaches: rational design and directed evolution[74,75]. Rational design requires basic knowledge of the structure-function relationship. This knowledge is utilized in the de novo synthesis of the desired biocatalyst[72,76−78]. With advancements in bioinformatics, an increase in the number of protein sequences, structures and computational tools such as HotSpot, Wizard, ProSAR, and SCHEMA have come about[72,78]. These tools can be utilized for analyzing the three-dimensional structures of enzymes, in order to alter, modify or design mutations for de novo enzyme design[78−81]. These data are evaluated to identify target amino acid residues and desired mutations that are then introduced into protein structures to manipulate biocatalysts. In one study, thermostable hydrolase from a thermophilic bacteria Thermobifida fusca (TfH) was isolated, characterized and expressed in recombinant Escherichia coli[30]. This enzyme was able to degrade the polyesters containing aromatic constituents and possesses the combined characteristics of lipases and esterases. Three-dimensional structure analysis of the enzyme showed that its structure differs significantly from usual lipases. The active site was shallow and protected compared to other lipases. The study suggested that with detailed knowledge of structure and protein design, this enzyme could be utilized for degrading and recycling PET or PBT on a industrial scale[30].
Functional enhancement of biocatalysts could be achieved by exploring the direct evolution of microbes under particular environmental conditions[80−82]. Direct evolution uses combinatorial methods to alter specific properties of enzymes to achieve hallmark improvements in enzyme activity for specific substrates[80]. Direct evolution involves subjecting the gene encoding the enzyme of interest to repeated rounds of mutagenesis to construct a library of variants. These variants are further screened and selected based on the desired function. The most improved single variant is then subjected to repeated cycles of mutagenesis and selection until no further improvement is shown by the variant in its desired property[82]. Recently, directed evolution of a bacterium Ideonella sakaiensis 201-F6, grown on PET has been reported[27]. The bacterium was grown on PET and found to produce two enzymes, PETase and MHETase, which can hydrolyze the PET polymer into its monomers; terephthalic acid and ethylene glycol. The ability to control and engineer biocatalyst parameters such as shape, rigidity, flexibility, composition and surface chemistry offers a catalog of possibilities for utilizing biocatalysts in biocatalyzed plastic degrading technology. These engineered biocatalysts can then be mass produced using transgenic microbes[83].
Biocatalyst availability and in situ product recovery
-
For economical large-scale application of biocatalysts, it is essential that they can be re-used. Enzyme immobilization provides an excellent basis for increasing availability of an enzyme to its substrate with greater turnover over a considerable period of time[84]. For immobilization, first a suitable non-reactive and stable matrix is selected which may be natural or synthetic in nature. The enzyme is immobilized onto the solid matrix using a suitable technique such as trapping, encapsulating, adsorption, covalent bonding or copolymerization[85,86]. In this form, the enzyme is readily available for its substrate. Generally, plastic is an insoluble material and in bulk amounts it floats on the surface of the tank, limiting its bioavailability to the biocatalyst[31]. Therefore, another widely used application of the immobilization approach together with enzymes has been an enzymatic reaction on immobilized substrates.
In enzyme-catalyzed reactions the product itself acts as a negative feedback inhibitor of the enzyme. Therefore, in situ product recovery is supposed to increase the efficiency of the overall process[86]. Such techniques could be integrated into the bioreactors for product recovery and recycling. The techniques for product recovery must be based on the degree of product enrichment, improved productivity, reduced process flows and increased yields[86]. Bi-phase extraction systems are valuable techniques used for product recovery in many biocatalytic reactions. At first, the biocatalytic reaction occurs in a homogeneous media, after which small changes in temperature induce the formation of two phases. This achieves the complete separation of the enzyme from the products in a single-step. The biphasic systems not only allow product recovery but also allow complete recovery of the enzyme – therefore facilitating its repeated use.
-
Biodegradation of plastics using microbial biocatalysts is a valuable technology. The microbial biodegradation of plastics at an acceptable level and efficiency is being enhanced. Designing and implementing natural or artificially created biocatalysts that can effectively degrade plastic on an industrial-scale to reduce plastic is needed. Discoveries of plastic degrading microbes and their enzymes opened up a completely new approach to plastic recycling and waste management. These discoveries could lead to the development of new methods to manage the billion tons of plastic accumulated globally. So far, biodegradation studies of plastic by microbes have been generally conducted on a laboratory scale by cultivating microbes in simple flasks. In order to scale up the process, several factors must be considered. For example, the growth rate of microbial cells might differ significantly between a shaken flask and an aerated bioreactor. Therefore, to scale up the process of biocatalyzed plastic degradation, it is recommended to use modified and controlled microbes that can grow rapidly to produce the maximum quantity of the specific biocatalyst. Moreover, an automated bioreactor with controls and biosensors based on computational models and software should be designed to build a user interface to control functions such as flow rate, stirrer speed, pumping, aeration, enzyme concentration, substrate concentrations, media volume, temperature and pH of the physical reactor setup. Modelling studies based on 13C-labelled plastic substrates should also be conducted to gain a better understanding of degradation mechanisms. This information should subsequently be used to build mathematical models of degradation prior to scaling-up degradation and plant-scale implementation.
This work was supported by Chinese Academy of Sciences, President’s International Fellowship Initiative (CAS-PIFI), Grant No. 2019PC0011. All the authors of the paper are thankful to Dr. Fiona Worthy for the English edition of the manuscript.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visithttps://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Khan S, Nadir S, Iqbal S, Xu J, Gui H, et al. 2022. Bio-catalyzed plastic degradation: a review. Circular Agricultural Systems 2:5 doi: 10.48130/CAS-2022-0005
Bio-catalyzed plastic degradation: a review
- Received: 11 April 2022
- Accepted: 12 July 2022
- Published online: 10 August 2022
Abstract: The widespread use and production of plastic have led to increased accumulation of plastic waste in the environment which threatens terrestrial and marine life. Efficient methods for management of plastic waste remain a key challenge. Biodegradation of plastics is considered an environmentally safe method, but is still limited to laboratory scale. Several previous studies have reported microbial enzymes capable of degrading plastic. These discoveries offer a promising starting point for the development of biocatalyzed plastic degradation technology. In this review, we discuss recent advancements and applications of biocatalyst technology. We also describe the different steps for development of bio-catalyzed plastic degradation technology and the major issues related to each stage. Breakthroughs in research into biocatalyzed plastic degradation would lead to new opportunities for sustainable alleviation of the worldwide problem of plastic waste accumulation.
-
Key words:
- Biodegradation /
- Enzymes /
- Microbes /
- Plastic /
- Recycling /
- Technology












