-
Plant tissue culture systems are fundamental tools for investigating gene functions in higher plant species. Plant tissue culture and Agrobacterium-mediated transformation have profoundly reshaped and promoted functional studies in crop research and breeding over the past three decades. Rice (Oryza sativa L.) is a staple food that feeds more than half of the world's population[1]. Since the end of the 20th century, protocols for tissue culture and Agrobacterium-mediated transformation in rice have been established[2−4]. Subsequently, scientists have expanded applications of these methods to allow use in more cultivars[5−8]. Several other transformation methods have also been applied to generate transgenic rice plants, including biolistic technology-based methods and nanotechnology gene delivery systems[9,10]. During the last century, particle acceleration, also known as microprojectile bombardment or gene gun technology, has been successfully applied to several commercial crops such as Roundup soybeans[11]. However, this technology has limitations; it induces cell damage and has low transformation efficiency and genome integration rates[12]. In recent years, nanoparticles and nanotechnologies have been widely applied in animal cells and organs. In this context, nanoparticles function as molecular vectors to harmlessly deliver bioactive materials into cells, and have therefore been utilized as targeted drugs, cancer treatments, and genetic disease cures[13]. However, this novel technology is far from ready for widespread application in plants.
Agrobacterium tumefaciens-mediated transformation has been used to transform rice for over 20 years[2], but species- and genotype-specific difficulties have hindered broad application. Moreover, although there are classic japonica cultivars (such as ZH11 and Nipponbare) that can be used for transformation, there are no standard genetically tractable indica cultivars. Significant progress has been achieved in developing transformation methods; for instance, some indica cultivars have been recognized as easier to transform, tissue culture components have been improved[8], the essential genes that determine callus browning and regeneration rates have been identified[14], and specific enhancements have been made to transformation protocols[15]. However, a 'gold standard' protocol has yet to be established for functional studies and other applications of rice transformation, particularly in indica cultivars.
Grain weight is one of the key factors determining single plant yield production of rice and other cereal crops[16]. Reports on trade-off between grain number and grain size emerged and focused on the famous OsMKKK10-OsMKK4-OsMPK6 cascade[17,18]. Grain Size and Number 1 (GSN1) encodes a dual-specificity phosphatase which negatively regulated the MAPK cascade and functioned as a developmental 'brake' in coordinating localized cell differentiation and proliferation[19]. Previously research confirmed that overexpression of GSN1 reduced grain size, while knockout or knockdown of it would significantly enlarge the grain size[19,20]. This gene also provided huge potentials on grain size and yield production improvement.
We previously found that a novel elite indica cultivar, Nan Gui Zhan (NGZ)[21] (bred by the Rice Research Institute, Guangdong Academy of Agricultural Science), is suitable for rice transformation assays. To better evaluate the transformation regeneration rate, we here used optimized protocols appropriate for the indica cultivar Zhen Shan 97 (ZS97) and performed a comparison of overexpression and CRISPR-Cas9-mediated gene editing plasmid transformations. We compared five distinct elite indica rice cultivars and one japonica cultivar from South China, Central China, Southeast China, and India. We generated GSN1 gene constructs; performed plant tissue culture experiments; measured callus induction and regeneration, root regeneration, and shoot regeneration rates; evaluated tissue culture performance; and assessed T2 generation phenotypes and genotypes. Finally, we analyzed several genes related to callus regeneration and the associated haplotypes and expression patterns in these cultivars, which partially explained the differences in tissue culture outcomes. Our results indicated that a popular cultivar of rice with high yield production and high grain quality is sufficient for Agrobacterium-mediated transformation, indicating that this cultivar has a promising future in gene editing applications and functional studies associated with indica rice breeding.
-
Five different indica rice cultivars (NGZ, ZS97, YD 6, HHZ, and Kasalath) and one rice japonica cultivar (ZH11) were obtained from BioRun Co. LTD. All grains were harvested in the same period in Hainan, 2020. All seeds were healthy and similar in size and appearance. All rice plants were cultivated in Guangzhou, China, under natural growth conditions.
Plasmid construction and plant transformation
-
To generate the overexpression construct pCAMBIA1300-GSN1-FLAG, the full length of GSN1 cDNA (LOC_Os05g02500) was cloned from rice elite cultivar FAZ1. The GSN1 coding sequence was fused to a 3× FLAG tag and cloned into a plant binary vector with the Maize ubiquitin (UBI) promoter. To produce the CRISPR-Cas9 gene editing rice, we applied the pCAMBIA1300-sgRNA-Cas9 system for plasmid construction and sgRNA design[22,23]. We constructed the pYLCRISPR/Cas9 Pubi-GSN1-H plasmid for gene editing transformation. Two gRNAs (GSN1-U3F: 5'-GGCAGTTCTGGCGCTCCGCGTCG-3'; GSN1-U3R: 5'-AAACCGACGCGGAGCGCCAGAAC-3'; GSN1-U6AF: 5'-GCCGGCGGAGGGGGTGTCGGCGG-3'; GSN1-U6AR: 5'-AAACCCGCCGACACCCCCTCCGC-3') were utilized and introduced into the CRISPR-Cas9 main backbone by BsaI accordingly[19]. These well-constructed plasmids were subsequently transformed into competent cells (E. coli, DH5α). Agrobacterium tumefaciens-mediated transformation of rice with strain EHA105 was applied as previously described methods[2].
Mature seed tissue culture and transformation
-
Mature seeds were harvested in Hainan, 2020. The whole tissue culture process was modified according to the method of Zhao et al.[24]. To dehull the mature rice seeds, the moldy surface and rotted embryo seeds were removed, then the healthy full seeds were left for tissue culture. The seeds were disinfected with 75% alcohol for 30 s, followed by washing three times (ddH2O). The clorox chemical disinfectant was performed for 30 min, and seeds were washed using sterile water, three times. The seeds were then soaked in sterile water for an hour. After the disinfection process, the seeds were transferred to the callus induction medium to induce callus until the regeneration process. After the acquisition of a suitable size [2−2.5 mm (Kasalath), 3 mm (NGZ, HHZ), 3.5−4 mm (ZS97 and YD 6)] of callus, the plasmid-contained Agrobacterium tumefaciens (EHA105 strain) were cultivated by YEB medium supplemented with kanamycin (50 mg/L). For co-cultivation, the callus and strains were scraped into the suspension culture and shaken for 20 min at 180 rpm. OD600 = 0.1−0.2. The infected callus was co-cultured 24 °C, 48 h under dark conditions. The selection was performed on a resting medium with 75 mg/L hygromycin. Two steps of selection process were applied. For ZH11, the pre-selection medium needs extra supplemented 35 mg/L hygromycin; the first stable selection medium contained 45 mg/L hygromycin; the second selection medium added 60 mg/L hygromycin. For indica rice selection process except YD 6, the pre-selection medium needs extra supplemented 15 mg/L hygromycin; the first stable selection medium contained 25 mg/L hygromycin; the second selection medium added 60 mg/L hygromycin. For YD 6, the pre-selection medium needs extra supplemented 5−10 mg/L hygromycin; the first stable selection medium contained 15−25 mg/L hygromycin; the second selection medium added 60 mg/L hygromycin. For the differentiation process, the dense, beige, and middle-size transformed callus were transferred to a differentiation medium (light 28 °C, 24 h) for a week. Green spots were observed in certain callus; 1 cm long young seedlings were grown in the medium for two weeks. The regeneration seedlings were transferred to the root induction medium. After ten days of cultivation (30 °C, 24 h), the regeneration root developed significantly. The young seedlings are now ready to be transferred to the natural environment. A simple diagram was exhibited the detailed steps and conditions for the plant tissue culture of rice in Fig. 1. The detailed parameters of all media through the tissue culture process were summarized in Supplemental Table S1. The total days of all cultivars transformation are listed in Supplemental Table S2.

Figure 1.
Diagram showing how rice tissue culture is used together with Agrobacterium-mediated transformation.
Validation of transgenic plants
-
All transgenic plants were cultivated in natural conditions in Guangzhou (China). The DNA was extracted from the young leaves of each regenerated plant using the DNA extraction protocol as described[25]. For the CRISPR-Cas9 gene editing plants, special primers to amplify sequences with the target sites were used (GSN1-F: 5’-CTCCTCTCACCCGTGACCTC-3’; GSN1-R: 5’-TGGCCGAGCCGGCGCCTTCC-3’); Hygromycin B phosphotransferase gene (HPT) primers (HPTF: 5’-CGCATAACAGCGGTCATTGACTGGAGC-3’; HPTR: 5’-GCTGGGGCGTCGGTTTCCACTATCCG-3’). For the transcriptional level evaluation, the gene-specific primers (GSN1-qRT-PCRF: 5’-CATCTAAAGGAGCTCGGCAATA-3’; GSN1-qRT-PCRR: 5’-CGAGCATTCAGCACTTCCA-3’) were used. Total RNA was extracted from leave tissues using Trizol reagent (Invitrogen). A reverse transcription assay was performed using ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO). The quantitative RT-PCR analysis was performed on the CFX96 Touch Real-Time PCR Detection System (BioRad). The data were analyzed using the 2−ΔΔCᴛ method, and the qRT-PCR regents were Fast Start Universal SYBR Green Master Mix with ROX (Roche)[26]. The Actin gene was used as the internal control. All analyses were repeated three times. All the primers used in this study are listed in Supplemental Table S3.
Calculation of callus induction, regeneration, selection, and transformation efficiency
-
A thousand healthy dehulled seeds were selected for plasmid transformation and tissue culture assays from each rice cultivar. The callus induction efficiency was calculated as the induction calli divided the total seeds percentage on the callus induction medium. The resistant cellus induction efficiency was counted by the number of resistant callus induced divided by the total induced calli. The green spots emerging rate was counted by the number of green spots divided by the total resistant calli. The shoot formation rate is calculated by the same way. The transformation efficiency indicated the positive hygromycin selected samples. The gene editing transformation efficiency was based on sequence results, and overexpression lines were evaluated by qRT-PCR data.
Phenotypic and statistical data analysis
-
The whole plant phenotypic data and the seed observation were acquired by digital camera and Stereo microscope (Leica). Student's t-test was used in data analysis. Diagram figure was performed by the BioRender Software.
Sanger sequencing and sequence comparison
-
Based on MSU 7.0 version rice annotation database, we searched and designed primers to amplify callus-related gene fragments of six rice cultivars. All targeted sequences were performed by Sanger sequencing, and Lasergene Seqman software was used for DNA fragment comparison.
-
Previous studies have indicated that transformation is much easier and has higher efficiency in japonica than indica rice cultivars[27,28]. Our group previously bred an ideal temperature-sensitive conventional indica cultivar, NGZ[21]. Here we used ZH11, the most popular and widely used japonica cultivar in transgenic rice research, for comparison[29]. ZH11 is a typical japonica cultivar with high native efficiency in Agrobacterium-mediated transformation[29−31]. We selected another five indica rice cultivars for tissue culture evaluation. These cultivars had a range of phenotypes and originated from various regions in South China, Central China, and India. All were mature cultivars that have been consumed for a relatively long time (ranging from the 1960s to 2010s). We compared grain size between these six samples (Fig. 2), and found that they were highly diverse in critical grain-related agronomic traits (Supplemental Table S4). NGZ contained higher alkali spreading value and brown rice rate, lower milled width, chalky grain rate, chalkiness area, and amylose content among these five indica cultivars. Therefore, NGZ is a typical high grain quality rice cultivar that is suitable for functional studies and improvement experiments related to grain size and shape. It has a very low chalky grain rate (1.5%) and chalkiness area (0.5%), and these attributes, along with the shape and milled performance, are desirable to consumers. NGZ also has relatively low amylose content and protein content, giving it superior flavor (Supplemental Table S4)[32,33]. Based on the grain performance (Fig. 2), NGZ appears to be an excellent novel elite rice cultivar for studying grain size and panicles.
Plasmid constructs and transgenic plant development
-
We previously reported that Grain Size and Number 1 (GSN1), which encodes the MAPK phosphatase OsMKP1, negatively regulates the MAPK cascade and controls trade-offs between grain size and number[19]. We here used two plasmids to generate transgenic plants and evaluate tissue culture performance (Supplemental Fig. S1): one for GSN1 overexpression and the other for GSN1 knockout via CRISPR-Cas9 gene editing (Supplemental Fig. S1). A functional gene editing system together with an overexpression system would enable most molecular mechanistic studies in rice. The components of the callus induction medium, resuspension medium, regeneration medium, selection medium, differentiation medium, and root induction medium have been optimized for japonica and indica rice cultivars (Supplemental Table S1).
Callus induction evaluation
-
We put all indica rice cultivars into some aspect of callus induction medium (CIM) to perform callus induction assays. The CIM used for ZH11 was slightly modified with respect to the ratio of sucrose, glucose, and phytohormone (Supplemental Table S1). One hundred healthy, intact grains were selected from each rice cultivar for each transformation event, and there were five biological replicates (500 seeds in total per cultivar). When seeds were cultured on CIM, more than 65% of total seeds were induced into embryogenic calli (Tables 1 & 2). We performed statistical analysis to display the survival rates of all plant tissue culture stages among six cultivars (Fig. 3). After growing on CIM for 14 d, we removed the endosperm and roots to obtain pure callus tissue for the next stage of tissue culture. The japonica cultivar ZH11 had the highest callus induction rate at 88.7%. In comparison, NGZ and ZS97 had similarly high calls induction rates (82%, NGZ; 88.3%, ZS97) (Tables 1 & 2). The Kasalath rice cultivar had the lowest callus induction rate (64.2%). We observed yellowish, compressed callus shapes in most indica cultivars and ZH11. Only very small calli were generated from Kasalath (Supplemental Fig. S2). In general, most indica cultivars formed calli earlier than ZH11 (Supplemental Table S5). Based on the phenotypical and statistical analysis as well as the callus induction rate, we admitted that the six cultivars ranked as follows: ZH11 (88.7%) > ZS97 (83.3%) > NGZ (82%) > YD 6 (77.6%) > HHZ (72.7%) > Kasalath (64.2%) (Fig. 3, Supplemental Fig. S2, Tables 1 & 2). These results confirmed that the indica cultivar NGZ had excellent callus induction capacity under commercial CIM conditions.
Table 1. Number of samples and survival rate percentage at each transformation step with the pYLCRISPR/Cas9Pubi-GSN1-H (GSN1-KO) plasmid in six rice cultivars.
Sample name Sample quantity Mature seeds Callus induction Resistant callus Green spot Shoot induction Transformation NGZ 1000 (100%) 820 (82%)** 111 (26.7%)** 34 (30.6%)n.s 24 (70.5%)n.s 24 (4.8%)n.s ZS97 1000 (100%) 833 (83.3%)* 271 (62.0%)* 41 (15.1%)n.s 26 (63.4%)n.s 26 (5.2%)n.s YD 6 1000 (100%) 776 (77.6%)*** 54 (14.5%)*** 12 (22.2%)* 6 (50%)* 6 (1.2%)* HHZ 1000 (100%) 727 (72.7%)*** 261 (71.1%)* 36 (13.7%)n.s 22 (61.1%)n.s 22 (4.4%)n.s Kasalath 1000 (100%) 642 (64.2%)*** 196 (62.2%)n.s 140 (71.4%)* 17 (12.1%)n.s 17 (3.4%)n.s ZH11 1000 (100%) 887 (88.7%) 189 (41.6%) 55 (29.1%) 24 (43.6%) 24 (4.8%) Data are given as the mean. n.s, no significance, * P < 0.05; ** P < 0.01; *** P < 0.001 compared with the corresponding ZH11 using Student’s t-test. The callus induction rate was caculated from the overall induced callus which was used for the whole studies. Table 2. Number of samples and survival rate percentage at each transformation step with the pCAMBIA1300-GSN1-FLAG (GSN1-OX) plasmid in six rice cultivars.
Sample name Sample quantity Mature seeds Callus induction Resistant callus Green spot Shoot induction Transformation NGZ 1000 (100%) 820 (82%)** 217 (53.5%)* 69 (31.8%)* 39 (56.5%)n.s 39 (7.8%)n.s ZS97 1000 (100%) 833 (83.3%)* 283 (71.5%)*** 56 (19.8%)* 52 (92.9%)n.s 52 (10.4%)n.s YD 6 1000 (100%) 776 (77.6%)*** 243 (60.14%)*** 60 (24.7%)* 17 (28.3%)n.s 17 (3.4%)n.s HHZ 1000 (100%) 727 (72.7%)*** 146 (40.6%)n.s 62 (42.5%)* 23 (37.1%)n.s 23 (4.6%)n.s Kasalath 1000 (100%) 642 (64.2%)*** 229 (45.8%)* 64 (27.9%)* 13 (20.3%)* 13 (2.6%)* ZH11 1000 (100%) 887 (88.7%) 160 (36.9%) 206 (161.5%) 37 (17.9%) 37 (7.4%) Data are given as the mean. n.s, no significance, * P < 0.05; ** P < 0.01; *** P < 0.001 compared with the corresponding ZH11 using Student’s t-test. The callus induction rate was caculated from the overall induced callus which was used for the whole studies. 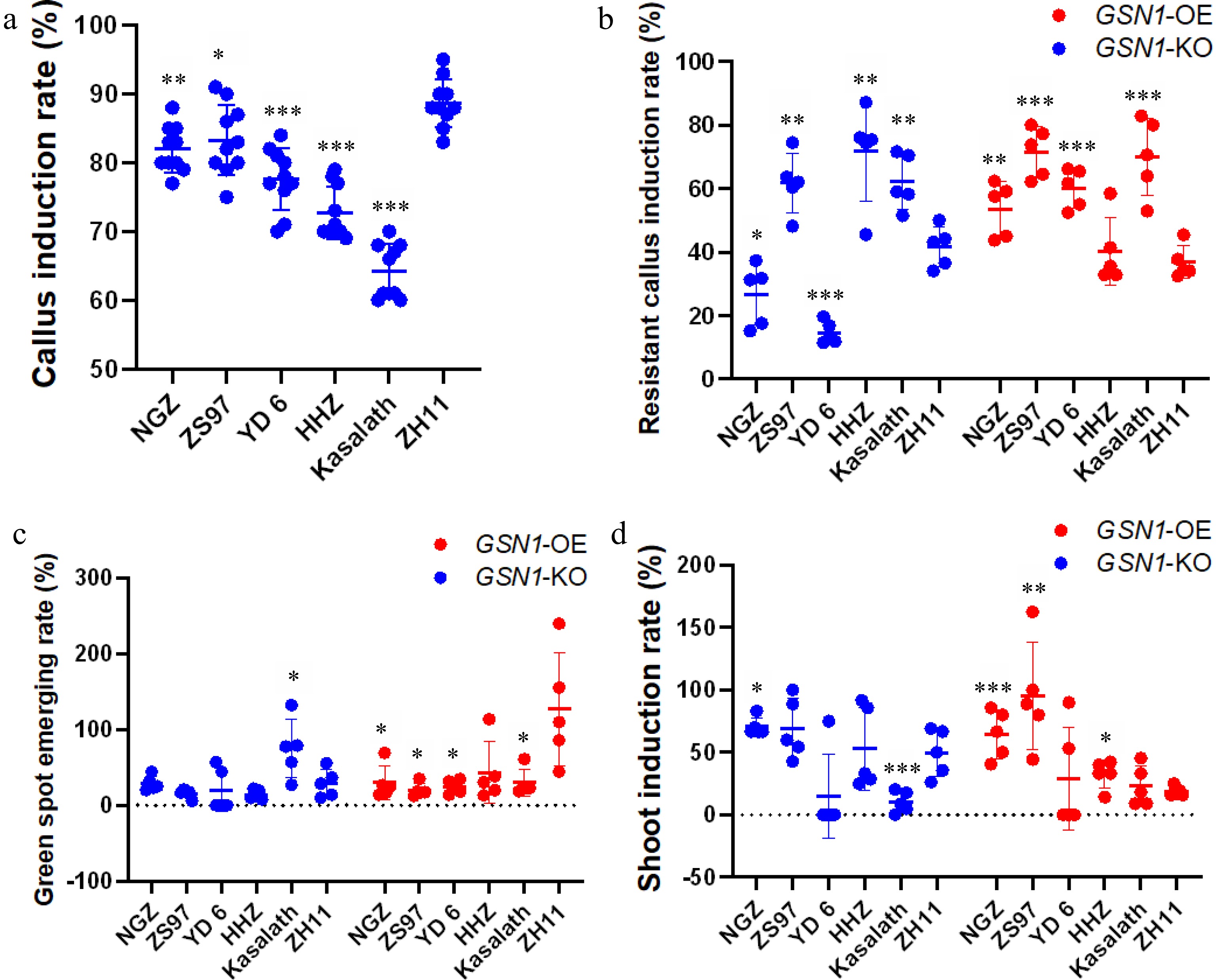
Figure 3.
Statistical analysis of plant tissue culture among six cultivars for the pYLCRISPR/Cas9Pubi-GSN1-H (GSN1-KO) plasmid and the pCAMBIA1300-GSN1-FLAG (GSN1-OX) plasmid. (a) The callus induction rate of six rice cultivars; (b) The resistant callus induction rate of six cultivars; (c) The green spot emerging rate of six cultivars; (d) The shoot induction rate of six cultivars. For callus induction process, ten independent biological repeats were performed (n = 10). For specific plasmid transformation process, five independent biological repeats were carried (n = 5), each performance utilized 100 healthy intact seeds from every cultivar for plant tissue culture process. Data are presented as the mean ± standard deviation. * < 0.05, ** < 0.01, *** < 0.001 (Student's t-test).
Survival rates of transformed resistant calli
-
We next transformed Agrobacterium tumefaciens strain EHA105 with either the overexpression plasmid pCAMBIA1300-GSN1-FLAG (GSN1-OX) or the gene editing knockout plasmid pYLCRISPR/Cas9Pubi-GSN1-H (GSN1-KO). All calli from each rice cultivar were incubated with transformed Agrobacterium in a co-cultivation medium under dark conditions at 28°C for one day. Calli were then transferred to resistant calli selection medium and grown for two weeks under dark conditions for the first round of selection. We observed that fresh calli of different cultivars differed in appearance and behavior. ZH11 calli were more yellow and compressed, had longer induction times, and generated multiple small fresh calli. Calli of the five indica cultivars were generally a lighter yellow (although some were white) and larger. The indica cultivars calli displayed severe browning or even death over the course of the cultivation time. We observed that in comparison to the other rice cultivars, NGZ was more sensitive to Agrobacterium infection (Tables 1 & 2). Statistical analysis showed that except HHZ, the overall resistant calli induction rate was lower for those transformed with the CRISPR plasmid than the overexpression plasmid (Fig. 3). NGZ and YD 6 had the two lowest resistant calli induction rates in GSN1-KO transformation event (Fig. 3). Many NGZ calli had Agrobacterium contamination; kanamycin was insufficient to inhibit agrobacterial growth. Successfully induced NGZ resistant calli were dry and clean (Supplemental Fig. S3). A similar phenomenon was observed in YD 6, but the percentage of contaminated calli was lower than the percentage of brown calli. ZS97 had the best performance, with a higher resistant calli induction rate than even ZH11 (Tables 1 & 2). Kasalath had similar responses to transformation with the two plasmids, whereas NGZ, HHZ and YD 6 had different responses to transformation with the gene editing and overexpression plasmids (Fig. 3). With respect to regeneration rate, the six cultivars ranked as follows: ZS97 (62%, 71.5%) > Kasalath (62.2%, 45.8%) > HHZ (71.1%, 40.6%) > ZH11 (41.6%, 36.9%) > NGZ (26.7%, 53.5%) > YD 6 (14.5%, 60.14%) (Tables 1&2). Overall, these results suggested that transformation with the gene editing plasmid led to a lower resistant calli induction rate, and that NGZ calli were sensitive to Agrobacterium infection.
Differentiation evaluation
-
After 2−3 weeks of regeneration growth, calli were transferred to differentiation medium and incubated at 28 °C under full-light conditions. Interestingly, indica cultivar Kasalath calli transformed with the GSN1-KO construct and japonica cultivar ZH11 calli transformed with the GSN1-OX construct showed large number of green spots (Supplemental Fig. S4). Consistent with previous reports, indica cultivars had lower greening numbers than japonica cultivars[34]. Although a considerable number of regenerated NGZ calli transformed with the GSN1-KO construct died, more than 30% of the calli exhibited their greening area. Kasalath had the highest greening rate at 71.4%. ZS97 and HHZ had the lowest percentage of greening (15.1% and 13.7%). With respect to greening rate, the six cultivars were ranked as follows: Kasalath (71.4%) > NGZ (30.6%) > ZH11 (29.1%) > YD 6 (22.2%) > ZS97 (15.1%) > HHZ (13.7%) (Table 1).
Calli transformed with the GSN1-OX construct had higher greening rates compared to those transformed with GSN1-KO. Surprisingly, ZH11 had a higher greening rate than calli regeneration rate. This phenomenon may be due to the high regeneration capacity of calli and the induction rate in japonica cultivars (Table 2). Furthermore, NGZ had the highest greening amount among all indica cultivars tested, indicating that NGZ had robust viability in plants and calli. Although the resistant callus induction rate was relatively low in HHZ, the greening rate was 42.5% for GSN1-OX transformants, suggesting that HHZ also had a strong capacity for greening (Table 2). We further investigated the size and shape of calli for all six cultivars. NGZ and ZS97 had larger calli and greening areas, whereas YD 6, HHZ, and Kasalath had green spots on smaller calli. Moreover, in comparison to ZH11, all indica cultivars showed darker calli and few were newly generated (Supplemental Fig. S4). With respect to differentiation, the six cultivars ranked as follows: ZH11 (161.5%) > HHZ (42.5%) > NGZ (31.8%) > Kasalath (27.9%) > YD 6 (24.9%) > ZS97 (19.8%) (Table 2).
Shoot and root growth evaluation
-
After 2−3 weeks of growth in differentiation medium, young plants were transferred into shoot induction medium (SIM). During this stage, there was a difference in shoot formation between GSN1-KO and GSN1-OX transformants (Tables 1 & 2). There were fewer GSN1-KO than GSN1-OX lines, and the shoot formation time was longer for GSN1-KO transformants. Although Kasalath plants showed more than 140 greening spots in total, only 17 transformed plants formed shoots (12.1%). Aside from Kasalath, the indica cultivars had higher shoot formation rates than the japonica cultivar ZH11 (Table 2). Similarly, in the GSN1-OX transformation assay, Kasalath had the lowest shoot induction rate (20.3%). Surprisingly, the ZH11 shoot induction rate in GSN1-OX (17.9%) was lower than the shoot induction rate with the GSN1-KO plasmids (43.6%) (Tables 1 & 2). ZS97 had the highest shoot formation rate (92.9%). Moreover, shoot and root lengths were longer in GSN1-OX than GSN1-KO transformants across all cultivars (Supplemental Fig. S5). Hyperhidrosis was not observed at any stage of the plant tissue transformation process. Plants that formed shoots and roots were selected with Hygromycin B. All shoot-forming plants counted in Tables 1 & 2 were confirmed as transformants. Notably, quite amount of transformed YD 6 calli and plants were killed by Hygromycin B in the shoot formation medium, consistent with previous research[35].
T1 transgenic plant phenotyping
-
The T0 seedlings of the six cultivars bearing the GSN1-OX and GSN1-KO plasmids were grown in Hainan (China). T1 seeds were harvested from T0 plants. Leaf pieces (2 cm) from each transformed plant were collected for Hygromycin B verification and PCR product amplification (Supplemental Fig. S6a and S6b). The results showed that all samples had been successfully transformed and revealed alterations at the molecular level. Sanger sequencing was conducted to verify the GSN1-KO constructs in all six cultivars (Supplemental Fig. S7); the results showed that all transformed lines had been successfully modified, functionally disabling GSN1 in those lines. Seed phenotypes were measured and analyzed in all wild-type and transgenic lines in the six cultivars (Fig. 4). The positive transgenic plants' phenotypes are well associated with genotypical alteration. We also performed qRT-PCR and confirmed that the GSN1-OX transformed plants in six cultivars have successfully increased the expression level of GSN1 (Supplemental Fig. S8). Two independent lines derived from each cultivar were selected for each construct to further investigate GSN1 function. Grain length was similar in all rice cultivars; the GSN1-OX and GSN1-KO lines had reduced and increased grain length, respectively. However, Kasalath and HHZ did not show significant grain elongation because the transgenic lines were heterozygous. ZS97, YD 6, and Kasalath showed decreased grain width in GSN1-OX lines, whereas GSN1-KO lines had wider grains. Interestingly, two high grain quality rice cultivars, NGZ and HHZ, showed different trends in seed width in GSN1-OX lines compared to the other cultivars. However, the GSN1-KO lines had wider grains in these cultivars, similar to the other cultivars. Grain thickness was similar between cultivars, except HHZ, for each type of transformant. In general, the GSN1-OX lines had thinner grains than the wild-type lines, whereas GSN1-KO lines had thicker grains. In contrast, HHZ showed the opposite effects in the OX and KO lines. In summary, transforming rice plants with a GSN1 gene editing knockout or overexpression construct led to changes in seed shape across all six rice cultivars, as previously reported[19].

Figure 4.
Phenotypes of T2 seeds of the six cultivars used in this study. For each cultivar, the average grain length (at left in pink, a1−f1), grain width (at middle left in blue, a2−f2), and grain thickness (at middle right in green, a3−f3) is shown for the wild-type, two GSN1-overexpression lines (OX-1 and OX-2), and two GSN1-knockout lines (KO-1 and KO-2). On the right (a4−f4) are grain phenotypes in the wild-type and mutant lines for each cultivar. Scale bar = 1 cm, n = 10.
Expression patterns and single nucleotide polymorphisms (SNPs) in potentially essential genes contributed to rice callus transformation efficiency
-
To further explore the molecular basis of differences in transformation efficiency between the selected indica and japonica cultivars, we performed quantitative reverse transcription PCR (qRT-PCR) to verify expression of callus formation-related genes and to establish the associations between gene expression patterns and cultivars (Fig. 5). The expression of our callus-related genes, including OsCRL, OsBOC1, OsBBM3, and OsSET1, were evaluated[14,36−38] (Fig. 5). The expression of OsBBM3 was significantly higher in callus samples from Kasalath compared to the other cultivars. Also, Kasalath owned the highest expression among all six cultivars of these four genes. The rest callus samples showed relatively lower expression levels (Fig. 5). We also found that HHZ has different expression patterns in OsBOC1, OsCRL and OsSET1 (Higher expression in OsCRL, but lower expression in OsBBM3, OsBOC1 and OsSET1). Except the OsBOC1, the expression of other three genes in YD 6 exhibited significantly lower than that in other cultivars. To further investigate the associations of haplotypes, expression levels and phenotypes, we performed Sanger sequencing to identify insertion/deletion mutations (InDels) and SNPs in these genes (Fig. 6). We cloned the promoter regions, untranslated regions (UTRs), coding regions, and introns of these genes. The sequencing results showed that two OsCRL SNPs were non-synonymous. N143D seemed to exist in japonica and indica, position 143 Asparagine (D) seems to be existed in japonica, and position 143 Aspartic acid (D) are shown in indica cultivars. In this gene, H114N is also another SNP that appeared in different cultivars. Position 114 Aspartic acid appeared only in NGZ and Kasalath (Fig. 6a). OsBOC1 encodes a SIMILAR TO RADICAL-INDUCED CELL DEATH ONE (SRO) protein that contributes to callus browning[14]. We identified two SNPs in coding regions, which is R32S and P377L. In our sequencing results, the R32S and P337L only existed in NGZ and Kasalath (Fig. 6b), the other rice cultivars did not show significant SNPs or InDels at that locus. We did not identify nonsynonymous SNPs or InDels in the coding regions of OsBBM3 or OsSET1 among the tested cultivars (Fig. 6c & d). In summary, our results revealed that the callus induction-related genes might contribute to the callus browning and induction rate variation in japonica and indica cultivars.

Figure 5.
Quantitative reverse transcription PCR (qRT-PCR) analysis of the callus related genes. Samples are harvested from calli after 7 d in CIM. The Actin gene was used as internal control to normalize gene expression data. Values are given as the mean ± SD. * P < 0.05; ** P < 0.01 and *** P < 0.001 compared with the ZH11 control using Student’s t-test. Standard deviations were calculated from three biological replicates.

Figure 6.
Single nucleotide polymorphisms (SNPs) and insertion/deletion mutations (InDels) in four genes related to plant tissue culture in seven cultivars. The key and main SNPs and InDels are shown in the promoter regions, untranslated regions (UTRs), coding regions, and introns. Genes were analyzed for the six cultivars included in the experimental section of this study using Nipponbare as the reference genome. Results are shown for (a) OsCRL, (b) OsBOC1, (c) OsBBM3, and (d) OsSET1. Bases shown in red in (a) and (b) indicate potential functional SNPs that may contribute to callus induction and browning.
-
This study compared transformation efficiency of several rice cultivars and identified genetic variations that may explain variations in transformation efficiency between cultivars. Transformation has been a challenge in indica cultivars for decades. Previous transformation methods have functioned well in japonica rice cultivars[5] such as ZH11 and Nipponbare. However, the cost and low transformation rate have hindered functional studies and application of indica cultivars. At present, optimized protocols have been developed only for japonica and a few indica cultivars[8,15,39]. Several strategies can be used to advance rice transformation methods and address this issue. First, more elite rice cultivar transformation optimization protocols should be invested in and developed. There is currently little incentive for biotechnology companies to invest in optimizing protocols without support and/or funding from research institutes, universities, or the government. Universities and companies are likely to only apply fully developed methods to novel rice transformation assays, which would not improve the transformation rate. Therefore, developing a specific project or increasing support for new method development would promote optimization of novel rice cultivar transformation methods. In addition, researchers should be encouraged to apply novel and advanced transformation methods. The field of plant biotechnology engineering currently uses biolistic technology and Agrobacterium-mediated transformation as the primary foreign material delivery methods in plants[9,40]. Within more than half a century of development, few species-specific transformation protocols have been established. The uncertainty of the integrated positions of foreign DNA fragments and Agrobacterium DNA contamination have decreased public trust in transgenic food safety[15]. However, the nanoparticle-mediated plasmid DNA delivery method has been successfully developed and applied to rice leaves and seeds[41,42]. This novel technology could aid in improving rice transformation methods.
NGZ, an elite indica cultivar, was developed and validated in 2016. Its grain quality is level I based on the Chinese Ministry of Agriculture and Rural Affairs Grain Quality Standard (GB/T17891-2017). The average grain yield of NGZ is over 8.25 t/hm2. This rice cultivar has been a main recommended cultivar for growth in South China since 2020. Plant tissue culture experiments in the present study showed that this cultivar has a high induction rate compared to other common indica species. Our data also showed that during the callus induction phase, 82% of calli had been generated by 14 d in NGZ, which was close to the rates of ZS97 and ZH11 (Tables 1 & 2 and Supplemental Fig. S2). However, NGZ had unexpectedly low resistant callus induction rate in both the GSN1-OX and GSN1-KO transgenic lines compared to the equivalent transformants in ZS97 and ZH11 (Tables 1, 2, Fig. 3, and Supplemental Fig. S3). In the latter stages, including the green spot, shoot, and root formation stages, NGZ performed as well as the ZS97 rice cultivar (Tables 1 & 2). Except for the Agrobacterium infection stage, NGZ generally had superior transformation and induction rates compared to the other indica cultivars. Interestingly, NGZ and the other indica subspecies used the same chemical reagents and protocols while ZS97 is well optimized (Supplemental Table S1). These results indicated that there could be considerable improvement in NGZ transformation efficiency.
We tested a reduced Agrobacterium co-culture time period to alleviate NGZ callus browning and contamination problems without much success (data not shown). We concluded that it is possible that the regeneration-related genes, such as OsBOC1 and OsSET1, expression mode is not suitable in NGZ when co-cultured with Agrobacterium. A new reports focusing on WOX11 and CRL1 (OsCRL) positively regulated root hair development, and it have shown that the importance of CRL genes in root development[43]. It is possible that the low regeneration efficiency in YD 6 is due to the low expression rate of CRL in it (Figs 5 & 6, Tables 1 & 2). Previous reports indicated that the upregulation of OsBOC1 is capable of reducing the browning rate in cultivated rice and wild rice[14]. In the present study, we found that the SNPs from Kasalath and NGZ are identical, indicating that the high callus transformation efficiency of OsBOC1 in NGZ is associated with Kasalath (Fig. 6). Interestingly, YD 6 callus is easier to be browned, but the qRT-PCR result of OsBOC1 in YD 6 sample is the highest among indica cultivars (Fig. 5), these results suggest that other factors might also be involved in YD 6 callus browning regulation. The expression of OsBOC1, OsCRL and OsSET1 are extremely high in Kasalath (Fig, 5, Tables 1 & 2), also hinted that the high transgenic successful rate in Kasalath. Recently, the maize GARP transcription factor gene GOLDEN2 was overexpressed in rice calli; this improved callus regeneration by activating chloroplast development[44]. In wheat, GROWTH-REGULATING FACTOR 4 (GRF4) and its cofactor GRF-INTERACTING FACTOR 1 (GIF1) function as a regulatory module and increase regeneration efficiency in transgenic plants[45]. Integrating the ZmBBM and GRF4-GIF1 modules into the CRISPR-Cas9 plasmid significantly enhanced the regeneration rate in maize[46], suggesting that these genes may function through a different pathway to regulate plant regeneration. It is possible that the GOLDEN2, OsBBMs, and GRF4-GIF1 modules would also increase the survival and regeneration rate in NGZ and other indica cultivars.
Except genetic factors, other physiological factors, such as Agrobacterium strains, phytohormone contents, carbohydrates, calli quality etc., also largely determined the plant tissue culture process and transformation efficiency[47]. Earlier studies focusing on improving specific rice variety transformation efficiency through plant tissue culture process. Optimizing this experiment by adjusting medium components as well as temperature, light density, agrobacterium concentration have successfully enhanced the transformation efficiency and resistant calli amount in certain rice cultivars[48]. This is also one of our future working directions to attempt linking physiological phenotypes to genetic basis, eventually we can optimize the rice transformation efficiency from the bottom.
In rice, GSN1 has been reported to control grain size and spikelet trade-offs[19], and ERECTA (ER) functions as an upstream regulator of the MAPK cascade to activate cytokinin signals during spikelet differentiation[49]. These regulatory networks contribute to spikelet formation and seed maturation. In this study, we used GSN1 as a case study to evaluate rice transformation efficiency (Fig. 4). Our data showed that overexpressing or knocking out GSN1 caused the largest phenotype changes in the indica cultivars HHZ and NGZ (Fig. 4). These typical high grain quality rice cultivars grow well in South China, and compared to other indica types, they had longer and thinner grains. However, overall yield production should still be increased. Our study provides potential strategies to improve gene editing and transgenic methods to achieve high yield in high grain quality rice.
Our work compared NGZ to the other four indica cultivars and a japonica cultivar in plant tissue culture regeneration efficiency and transformed efficiency. Although NGZ’s performance is not as excellent as ZS97 in indica cultivars (Tables 1 & 2), its good performance in plant tissue culture regeneration efficiency is recommendable. All media used in our experiments are called LY media, which is provided by BioRun Co. LTD. We listed all the ingredients and usages in Supplemental Table S1. According to technicians from BioRun Company, these media for rice indica cultivar plant tissue culture is well optimized, especially worked well in ZS97. However, ZS97 is an indica bred more than 40 years ago, its agronomical traits are not the best options for current rice studies. Although this is a well-optimized indica cultivar and performance the best in our experiment, NGZ, a newly bred indica cultivar in 2016, its grain quality, grain shape, abiotic stress, biotic resistance, and other essential agronomical traits are better than ZS97 and more suitable for phenotypical observation and studies in rice science. Moreover, NGZ’s plant tissue culture optimization and plant tissue culture potentials will be our effort in the following research aim. Thus, we hope that NGZ would be applied in gene editing research and molecular basis studies.
-
In summary, we here used GSN1 as a case study to compare the regeneration efficiency of one japonica and five indica rice cultivars. Each cultivar was transformed with a GSN1-overexpression plasmid and a CRISPR-Cas9 GSN1-knockout plasmid. Plants transformed with the gene editing plasmid had lower overall regeneration rates than those with the overexpression plasmid. NGZ and ZS97 had the highest transformation efficiency of the indica cultivars, but both required longer culture times than the japonica cultivar. We also verified expression patterns, SNPs and InDels of selected genes related to callus regeneration among these cultivars and provided possible explanations for differences in transformation efficiency between indica cultivars. Finally, we discussed the disadvantages of NGZ callus browning. We also discussed the advantages and possibility to apply NGZ in the future gene editing and functional studies. Our findings related to NGZ callus regeneration and reducing callus browning are expected to increase the application of NGZ in rice transformation assays.
We thank the BioRun Biotechnology Inc. Ltd. to perform the transgenic and plant tissue culture experiments. This work was supported by Laboratory of Lingnan Modern Agriculture Project (NZ2021001), Natural Science Foundation of Guangdong Province, China (2021A1515012160), Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science (R2021YJ-YB3015,R2019PY-QF007) and Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Ke Chen, Chanjuan Ye, Jie Guo
- Supplemental Figure S1 Plasmid construction diagrams used for this study.
- Supplemental Figure S2 Phenotypes of embryogenic calli from six rice cultivars in callus induction medium. Calli were induced in medium for six days. Scale bars = 1 cm and 0.2 cm, respectively. Statistical analysis is shown in Figure 3.
- Supplemental Figure S3 Phenotypes of embryogenic calli from six rice cultivars in resistant callus selection medium. Calli were incubated with Agrobacterium tumefaciens for 17 d for transformation. Scale bars = 1 cm and 0.2 cm, respectively. Statistical analysis is shown in Figure 3.
- Supplemental Figure S4 Phenotypes of embryogenic calli from six rice cultivars in differentiation medium. Green spots were observed in differentiation medium from eight to 19 d. Scale bars = 1 cm and 0.2 cm, respectively. Statistical analysis is shown in Figure 3.
- Supplemental Figure S5 Phenotypes of regenerated shoots from six rice cultivars in shoot induction medium (SIM). Shoots and young roots were observed in SIM. Scale bars = 1 cm and 0.2 cm, respectively. Statistical analysis is shown in Figure 3.
- Supplemental Figure S6 PCR verification and leaf verification confirmed that all transformed plants are hygromycin B positive plants.
- Supplemental Figure S7 The gene editing sequence results of GSN1-KO transgenic plants among six cultivars. Based on the construction map, two sgRNAs were designed and we found that both target sites were edited by CRISPR-Cas9 system. All kinds of base pair editing could be observed in our transgenic plants by Sanger sequencing.
- Supplemental Figure S8 The gene expression levels of GSN1-OX transgenic plants among six cultivars. The Actin gene was used as internal control to normalize gene expression data. Values are given as the mean±SD. *P<0.05; **P<0.01 and ***P<0.001 compared with the corresponding wild type control using Student’s t-test. Standard deviations were calculated from three biological replicates.
- Supplemental Table S1 The parameters of medium used for this study.
- Supplemental Table S2 The overall tissue culture time for each rice cultivar in this study. The total days counted from the seed disinfection day to all young seedling transferred to natural environment.
- Supplemental Table S3 Primers used in this study.
- Supplemental Table S4 Key grain-related traits among five indica cultivars. Mean values were shown in the table. Asterisks are marked as the significant perferable selection of grain quality. Data was collected by experiments and cultivar validation from IRRI and China Rice Data Center.
- Supplemental Table S5 The detailed plant tissue culture time for each rice cultivar in this study.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Chen K, Ye C, Guo J, Chen D, Guo T, et al. 2023. Agrobacterium-mediated transformation efficiency and grain phenotypes in six indica and japonica rice cultivars. Seed Biology 2:4 doi: 10.48130/SeedBio-2023-0004
Agrobacterium-mediated transformation efficiency and grain phenotypes in six indica and japonica rice cultivars
- Received: 20 December 2022
- Accepted: 30 January 2023
- Published online: 30 March 2023
Abstract: Plant transformation and regeneration have been continuously developed over the past four decades. In rice (Oryza sativa L.), Agrobacterium-mediated transformation has high efficiency in japonica and some indica cultivars using mature seeds and immature embryos. However, these protocols have low transformation efficiency in the latest indica cultivar that has been developed in South China since 2010. Here, we explored plant culture regeneration of the high-quality and high-yield indica cultivar Nanguizhan (NGZ) through CRISPR/Cas-based genome editing and traditional overexpression transformations. We compared transformation efficiency in this cultivar to four other widely-used indica cultivars and one japonica cultivar using mature seeds and the gene Grain Size and Number 1 (GSN1) as a case study. We observed universally smaller grain size in overexpression lines and bigger grain size in gene-editing lines among different cultivars. NGZ phenotypes make it an excellent model in which to investigate gene functions. We also examined the single nucleotide polymorphism (SNP) distribution and differences in expression levels of regeneration-related genes in calli, possibly revealing the source of NGZ's advantages in Agrobacterium-mediated transformation. These results shed light on the advanced application of NGZ in gene editing and overexpression transformation related to grain improvement, contributing to the 'rice breeding 4.0 era'.
-
Key words:
- Rice /
- Plant tissue culture /
- Agrobacterium-mediated transformation /
- Grain size /
- CRISPR-Cas9















