-
Plant defense response against invading pathogens is essentially chemical warfare occurring in nature. The plant cell can generate an enormous number of secondary metabolites with extreme structural diversity, in addition to primary metabolites including carbohydrates, lipids and proteins[1,2]. The major groups of secondary metabolites include phenols, terpenoids and alkaloids[3,4]. Secondary metabolites, including anti-microbial phytoalexins, anti-herbivory compounds, antioxidants, ultra-violet (UV) protectants, pigments, and aroma compounds, play important roles during plant interactions with the environment[5,6]. A wide variety of phenolic compounds such as monolignols, flavonoids, and hydroxycinnamic acid conjugates are produced from the phenylpropanoid pathway[1,7]. Notably, a large portion of the carbon in the phenylpropanoid pathway is channeled toward the synthesis of lignin, the second most abundant biopolymer eclipsed only by cellulose[3,7,8]. Many intermediates and end products of this pathway are vital to plant fitness, which allow plants to react fast and flexibly to various environmental cues including biotic or abiotic stresses.
Lignin formation is the outcome of oxidative polymerization of three p-hydroxycinnamyl (p-coumaryl, -coniferyl and -sinapyl) alcohols, or monolignols, which is catalyzed by both laccases and peroxidases[9]. Lignin composition can vary at the ratio or sequences of these monolignols, between plant taxa type of tissue, developmental stage, or environmental conditions[9,10]. These heterogeneous phenolic polymers are covalently cross-linked with cell wall polysaccharides which makes the cell walls structurally fortified and rigid as well as more resistant to microbial degradation[11,12]. The crucial role of lignin deposition in response to pathogen infection was proposed several decades ago[11,12]. More recently, accumulated molecular and cellular evidence from several pathosystems supports the notion that a lignified cell wall can serve as a physical barrier restricting phytopathogen intrusion[13−16]. Nevertheless, as cell wall lignification is a non-reversible process, tight regulation is expected over the induced lignification process including monolignol biosynthesis, polymerization and lignin deposition[17−19]. Until now the potential contribution of induced lignin formation as a primary component of apple root resistance to P. ultimum infection has only been suggested by available transcriptome data.
Biosynthesis of lignin and its detection in plant tissues has become a research hotspot recently, largely driven by the desire to reduce the recalcitrance of renewable lignocellulosic biomass[20−22]. Lignin detection has been traditionally targeted for evaluating wood characteristics in intensely lignified tissues. In contrast, fewer reports were related to detecting lignin in parenchyma cells of non-lignified tissues under pathogenic pressure such as in young apple roots[23]. To examine the potential role of induced lignin deposition in apple root resistance to P. ultimum infection, the methods need to be developed and tested to detect lignin deposition in young apple root tissue. Interestingly, several groups of molecules in plant tissues are known to be autofluorescent including two well-studied examples, i.e., chlorophyll (orange/red fluorescence) and lignin (blue/green fluorescence)[24,25]. Structural components in lignin molecules, such as phenolic rings and conjugated double bonds, are the important organic fluorophores[24,25]. Recently, we have explored the feasibility of assessing lignin deposition in young apple roots using fixation-free and label-free fluorescence imaging, in addition to brightfield observation based on histochemical staining of lignin[26].
Based on the findings from our recent transcriptome analyses that enhanced cell wall fortification from induced lignin biosynthesis may play a crucial role in defense activation in apple root. The current study aimed to further develop methods for assessing the lignin deposition patterns in apple roots between genotypes and/or in response to P. ultimum infection. Detecting the genotype-specific patterns of lignin deposition in young apple roots is crucially important for validating the functional roles of these candidate genes and defining their contribution to the observed resistance phenotypes. These methodologies of detecting lignin deposition patterns is vital for connecting transcriptome data of defense activation in apple root with the observed resistance phenotypes to P. ultimum infection.
-
The plant materials used in this study are apple rootstock genotypes from a cross between two elite apple rootstock varieties, 'Ottawa 3' and 'Robusta 5' (O3R5 for simplicity). Both parents have a strong background of wild apple germplasm[27,28]. Based on the recent phenotyping study, O3R5-#164 is resistant, while O3R5-#132 is susceptible to P. ultimum infection[29−31]. Tissue culture-based micro-propagation procedures were used to generate apple plants for infection assays as described previously[32]. Briefly, a four-week period of root induction in tissue culture medium was followed by one week of root acclimation in soil in a growth chamber before P. ultimum inoculation. To minimize transplanting shock as plants were transferred from tissue culture medium to soil, pots were placed within a 10-inch × 20-inch flat tray and covered by a transparent 7-inch Vented Humidity Dome (Greenhouse Megastore, Danville, IL, USA). Plants for tested genotypes were generated in the same schedule to synchronize the plant developmental progression. All plants were watered every other day and maintained under the same condition before or after pathogen infection.
Inoculation of apple roots with Pythium ultimum
-
The P. ultimum strain (#1062) used in this study was originally isolated from the roots of 'Gala'/M.26 apple grown at Moxee, WA, USA. Inoculum of P. ultimum was prepared as previously described[33]. Briefly, P. ultimum was cultivated in potato-carrot broth 20 g of carrots and 20 g of peeled potatoes in 1 L of water boiled for 30 min, with two drops of wheat germ oil added per litre of medium. The broth cultures of P. ultimum were grown in 9-cm petri dishes at 22 °C for 4−6 weeks. Oospores and mycelium from the resultant hyphae mats were collected and ground in sterile water using a household electric blender for 30 s. The oospores and hyphal fragments were resuspended in 2% methyl cellulose to give a final concentration of approximately 2,000 oospores per mL. The inoculation of apple roots with P. ultimum was performed by dipping the root system in the inoculum solution for 5 s. Inoculated plants were immediately transplanted into pots with autoclaved soil medium. Control plants were mock inoculated with 2% methyl cellulose solution and maintained under conditions similarly to the inoculated plants in an environmental growth room. Plant roots were sampled at 48 hpi (hour post inoculation) for sectioning, staining and microscope observation.
Lignin staining of sectioned apple root tissue and microscope observation
-
Root sections of about 30 to 50 µm in thickness were made using Personna #74-0002 razor blades. From each plant, many sections were cut from one root; the sections were then allocated randomly into three groups for two staining methods or no-staining controls, respectively. Hand-sectioned root tissue was stained using two histochemical staining methods reported to be specific to lignin: Wiesner (phloroglucinol-HCl) staining and Maule staining[23,34]. For the Wiesner test, sections were immersed in a solution consisting of 0.3 g phloroglucin, 10 ml ethanol, and 5 ml concentrated HCl for 10 min, then mounted in water on a slide. For Maule staining, sections were immersed in 0.1% KMnO4 for 5 min, rinsed with water, and mounted in 30% ammonium hydroxide. While Maule staining typically includes treatment with hydrochloric acid between the potassium permanganate and ammonium hydroxide steps[35,36], we omitted HCl treatment after observing that it usually led to disintegration of the root sections when coverslipped, and that exclusion of HCl treatment did not produce a visible difference in staining in our young root samples. Lignin deposition patterns were observed with brightfield and fluorescence imaging immediately after staining. For better comparability, the root segments selected for sectioning were located at a similar position of 1 cm from the root tip; the resulting root tissue sections represent similar, or comparable stages in terms of development and differentiation. Sections were visualized and documented using an Echo Revolve fluorescence microscope (Discovery Echo, San Diego, USA). For fluorescence imagery, the microscope was equipped with a 470/40 nm excitation filter, 525/50 nm emission filter, and 495 nm dichroic mirror. Microscope images were recorded for at least two representative root sections per plant within each stain treatment. A minimum of three plants were selected for sectioning and staining. Software parameters included low gain, exposure time 500 ms, histogram on Automatic, and LED power at 100%.
Flavonoids staining of sectioned apple root and stem tissue
-
Apple root sections were decolorized in absolute ethanol for 3 h and stained for the presence of proanthocyanidins and flavan-3-ols using 0.1% (w/v) 4-dimethylaminocinnamaldehyde (DMACA) in absolute ethanol containing 0.8% w/v hydrochloric acid. To answer the question of if proanthocyanidins are a major component in root tissue of apple, the stem sections were used as a positive control for detecting the presence of proanthocyanidins. The stained sections from both roots and stems were subjected for brightfield imaging using Echo Revolve fluorescence Microscope (Discovery Echo, San Diego, USA).
-
Brightfield images from two lignin-specific staining methods showed variable degrees of lignin deposition between genotypes and treatments (Fig. 1). The Wiesner test produced the typical pink coloration of lignin in select tissue types or cellular locations, especially at the highly enriched area around the vascular cylinder, or specifically the phi cell layer (Fig. 1, upper two rows, blue arrows)[37]. Between two genotypes, contrasting patterns of lignin deposition were observed immediately outside the vascular bundle based on Wiesner test (blue arrows, Fig. 1, top two rows). A more organized ring with the typical pink color can be easily detected in root sections from a resistant genotype #164. In contrast, a less defined pattern was exhibited at the same location in root sections from a susceptible genotype #132. However, induced lignin deposition from P. ultimum infection was not always discernable by Wiesner test between two genotypes. On the other hand, Maule staining generated the characteristic red to brown coloration of lignin from sections of young apple root (Fig. 1, lower two rows). It appeared that Maule staining is capable of detecting lignin more readily and with enhanced sensitivity, even in the parenchyma cells which normally accumulate relative low levels of lignin in their cell walls. Elevated lignin deposition was detectable in sections from P. ultimum infected root tissues stained with Maule reagent (Fig. 1, images on the right side of both two lower panels), as compared to those stained sections from non-infected control roots. Also interesting is the fact that sections from infected roots of the susceptible #132 are often less intact likely due to the brittle nature of infected tissue at 48 hpi. The preliminary conclusion is that the Maule test is more efficient in detecting lignin deposition in young apple roots, particularly for the parenchyma cells in outer cortex tissue (in addition to vascular tissue). However, the Wiesner test was able to reveal the contrasting patterns of lignin deposition in the phi cell layer. Therefore, both methods offered advantages for various aspects of detecting lignin deposition in young apple roots.
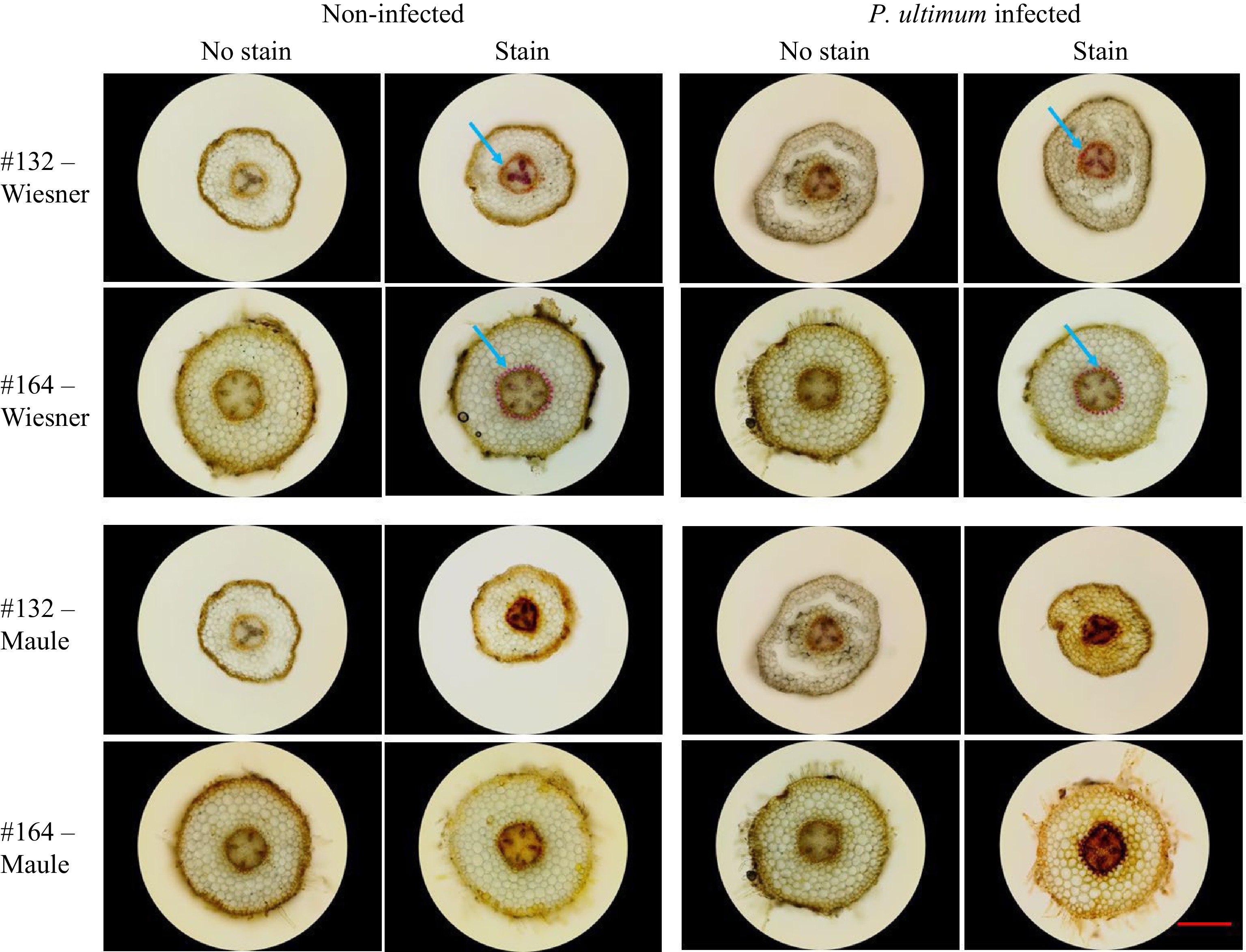
Figure 1.
Brightfield images of detected lignin deposition in apple roots. The top two rows are images from the Wiesner test, and the bottom two rows are from the Maule test. Two apple rootstock genotypes were included: the susceptible #132 and the resistant #164 each of which was infected vs mock-infected with P. ultimum. Each image is a representative of roots from three plants. The red bar represents 500 µm.
Fluorescence images of lignin deposition in apple roots
-
Fluorescence images in Fig. 2 revealed additional features related to the lignin deposition in apple roots. The same sections from which the brightfield images were collected were also subjected to fluorescent imaging. Relative to unstained control sections where autofluorescence were readily detected in most tissue types, lignin staining resulted in reduced autofluorescence emission[26]. Interestingly, two chemical staining methods exhibited differential quenching effects (yellow arrows). Wiesner reagent demonstrated a more dramatic effect on fluorescence quenching, while the fluorescence-suppressing effect is less obvious for the Maule test. Additionally, it appeared that the quenching effect due to staining was more obvious in the parenchyma cells than in the vascular bundles. Between genotypes, fluorescence from the resistant genotype #164 was less suppressed while the quenching effect is more easily discernable for the susceptible genotype of #132. Such discrepancy in fluorescence quenching between genotypes may reflect the levels and/or the composition of lignin in these root regions. Similar to the observations from brightfield images, the root sections of the resistant #164 appeared to contain more intense lignin deposition in the region immediate outside the vascular bundles compared to the susceptible #132, based on the detected fluorescence intensity (red arrows as examples). Within each genotype-by-treatment group, the fluorescence images of sections appeared to corroborate the corresponding brightfield images. Because of the specificity of both lignin staining methods, the quenching effect due to these staining reagents (yellow arrows) appeared to confirm that the observed fluorescence is primarily from lignin in the cell walls of young apple root tissue, rather than from other types of autofluorescent compounds. Additionally, it appears unlikely that the Wiesner and Maule reagents directly interfered with our excitation or emission light. Phloroglucinol shows virtually no absorbance near 470 or 525 nm, and although potassium permanganate does have an absorption peak near 525 nm, such interference should reduce fluorescence uniformly across the entire image, which was not the case for either stain technique.
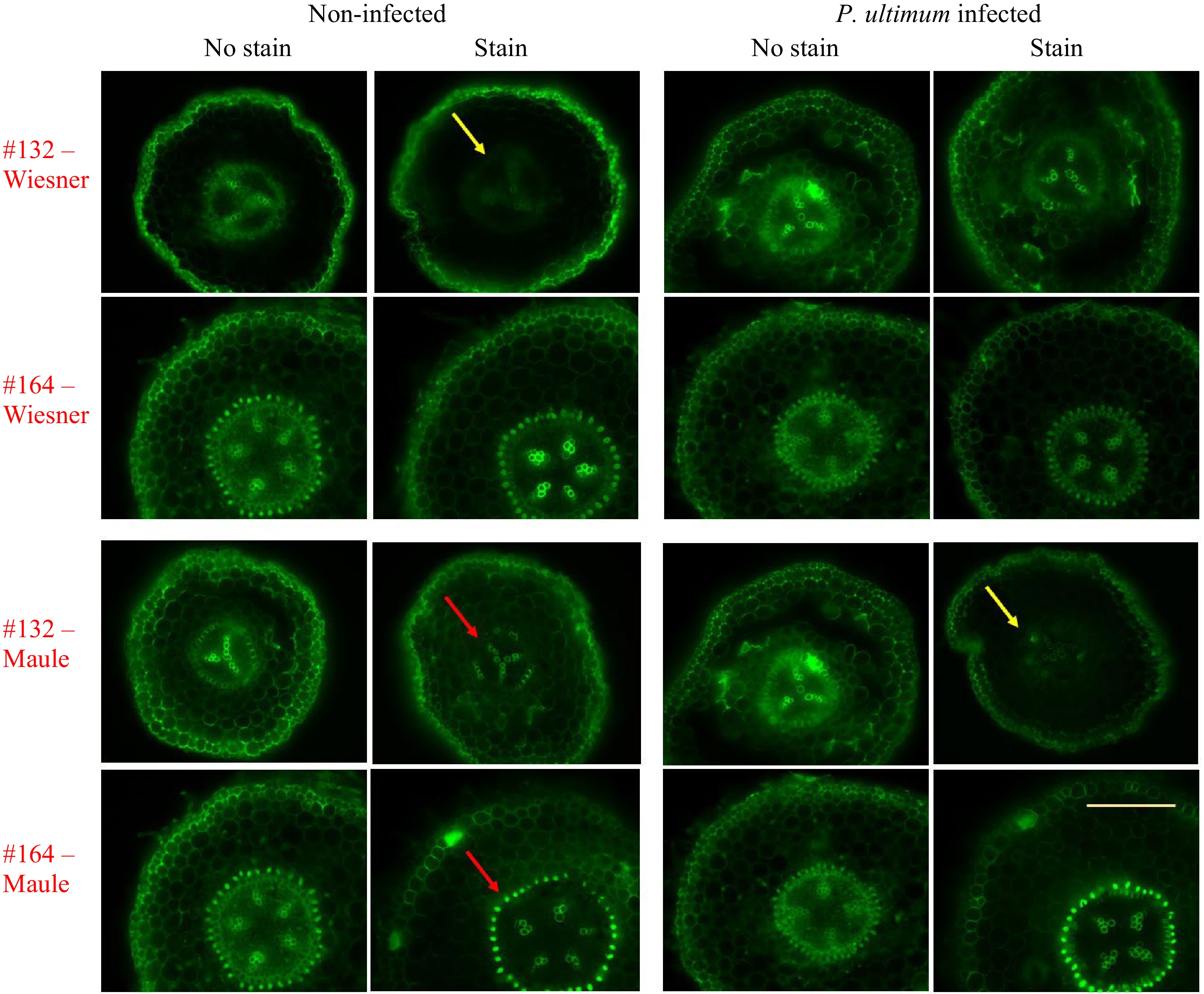
Figure 2.
Fluorescence images indicating lignin deposition in apple roots. The top two rows show sections from the Wiesner test, and the bottom two rows are those sections from the Maule test. Two apple rootstock genotypes were included: the susceptible #132 and the resistant #164. Each image is a representative of roots from three plants. The yellow bar represents 500 µm. Ex. 470/40 nm, Em. 525/50 nm, Dm. 495 nm.
DMACA staining of proanthocyanidin in cell walls of apple root tissues
-
Proanthocyanidins, or condensed tannins, are polymers of flavan-3-ol subunits, which are produced by the flavonoid biosynthesis pathway in many plants[38]. The presence of proanthocyanidins is often detected by histochemical staining using 4-dimethylaminocinnamaldehyde (DMACA)[38]. Using DMACA staining of root sections, no typical blue coloration from stained proanthocyanidins was detected in apple roots (Fig. 3, upper panel). As a positive control, leaf lamina from the same apple plant showed easily detectable blue coloration of proanthocyanidins (Fig. 3, lower panel). The result seemed to indicate that pro-anthocyanidin, which is autofluorescent as lignin, is not a major component in apple roots.

Figure 3.
DMACA staining of proanthocyanidin in cell walls of apple root and stem tissues. Top row shows sections from root tissue from #164. Absence of blue coloration indicates lack of proanthocyanidin accumulation in apple root tissues. Bottom shows two sections from apple leaf tissue. The blue color (red arrows) is present on both sides of the leaf as well as inside the main vascular bundle, suggesting the existence of proanthocyanidin in these locations. White bars represent 500 µm.
-
Development and deployment of resistant or tolerant apple rootstocks may provide a durable, cost-effective and environment friendly approach for managing apple replant disease, which is incited by a soilborne pathogen complex including the necrotrophic P. ultimum[29,39]. Conventional breeding for resistant apple rootstocks is a long-term and resource-demanding endeavor. Application of molecular tools can facilitate the breeding process, but a better understanding of resistance mechanisms in apple roots is the first step for developing these tools[29,39]. For any pathosystem, the host plant, or even different tissue types, could utilize a unique set of defense strategies to thwart the pathogen attacks and minimize damage to the host cells. To unravel the transcriptional network regulating defense activation in apple roots in response to P. ultimum infection, a series of transcriptome analyses have identified candidate genes associated with several pathways or processes which may directly impact the resistance traits[33,40−42]. Based on the analyses of transcriptome data, cell wall lignification appeared to be one of the most prominent components contributing to an effective defense strategy in apple root against infection from P. ultimum[42,43].
Cell wall lignification under pathogenic pressure has long been proposed to function as a physical barrier to restrict pathogen growth into tissues and limit the diffusion of pathogen-derived enzymes and toxins into cells[12,15]. Furthermore, low-molecular-weight phenolic precursors for lignin formation and free radicals produced during polymerization may inactivate enzymes, toxins, and elicitors from pathogens[12]. Lignin possess an interesting feature of autofluorescence emission, while polysaccharides are known to be non-fluorescent[44]. Although lignin is not the only autofluorescent compound in plant cell walls[24], its specific staining and the resulted quenching effect on fluorescence intensity can be a powerful evidence that the observed fluorescence is primarily from lignin in cell walls of young apple root. Furthermore, the failure to detect proanthocyanin (a dominant form of flavonoid) could further support the idea that lignin, rather than other phenolics from an upregulated phenylpropanoid and flavonoid biosynthesis pathways, is a major component of the defense response to P. ultimum infection. More-definitive evidence regarding the role of induced lignin deposition in disease resistance in apple root should come from biochemical quantification of lignin and transgenic manipulation experiments.
Several challenges exist in detecting induced lignin deposition during defense response in apple root, even though lignin is one of the most abundant biopolymers. For example, the elevated lignin biosynthesis in response to pathogen infection could be minimal in comparison to the abundant presence of lignin as a major structural component. Second, the induced lignin biosynthesis under pathogenic pressure might be specific to certain tissue types, at specific timing or certain subcellular locations. Specific to this pathosystem, as P. ultimum is a fast-growing filamentous pathogen, it is difficult, if not impossible, to determine the timing of pathogen infection at a specific root segment. In other words, after initial inoculation, the actual infection process at a specific location within the root system is difficult to track. The continuous root growth process, spontaneous lateral root initiation and high plasticity to surrounding conditions can also contribute to non-synchronized differentiation progression between root branches within a root system or between plants and/or genotypes. Therefore, sampling root segments with comparable physiological and/or pathological status is extremely challenging. Another practical challenge is the constant supply of root tissue for repeated assays, due to the destructive nature of the pathogen infection assay. Seed germination cannot produce so-called 'true to type' apple plants, due to the fact that apple reproduction is self-incompatible (or outcrossing in nature) and the apple genome has high-level heterozygosity[45,46]. To overcome this hurdle, a tedious and time-consuming tissue culture procedure must be implemented for the continuous availability of plants, or root tissues, for tested genotypes.
Detecting the microscopic patterns of lignin deposition is a key part of in-depth phenotyping of apple root resistance at the cellular and biochemical levels. Such data are essential for assigning the functional roles of specific candidate genes identified from transcriptome analysis and understanding their contributions to the observed resistance traits. Because of the abovementioned hurdles in detecting apple root lignin deposition patterns, a more rigorous standard for sampling specific root segments is pivotal for a reliable assessment of root defense responses such as lignin deposition patterns. Many parameters need to be further scrutinized for a more-detailed evaluation of the role of lignin to apple root resistance. With all these potentially limiting factors in mind, our preliminary data provides an overall assessment of lignin deposition patterns in apple root between genotypes and under the pressure of P. ultimum infection.
In the current study, several measures were taken to minimize variation in physiological or pathological status of selected root segments. For example, the root segments for sectioning were selected at the same distance from root tips, with the assumption that cellular physiology and differentiation phase would be more comparable. Root segments with similar diameters were selected from control and infected plants. Care was taken to minimize abiotic stresses by applying consistent growing conditions and handling procedures. Additionally, the thickness of hand-cut root sections was estimated using the Z-stack function associated with the ECHO microscope to minimize the differences of observed intensity in fluorescence or color from variable section thickness. It is worth noting that variations of lignin structure and composition as well as the linkages between lignin and carbohydrates can also impact the intensity of lignin fluorescence[25]. Minimizing irregularities in tissue sampling, sectioning, and imaging processes is a critical yet challenging issue for microscopic analysis of lignin deposition patterns in apple root tissues.
In summary, our preliminary observations indicated that when using both brightfield and fluorescence images, elevated lignin deposition was detected in the parenchyma cells of infected root cortex tissues in response to P. ultimum infection. Compared to Wiesner staining, the Maule test is more effective for detecting lignin in the less-lignified tissue in young apple root. The genotype-specific lignin deposition patterns, particularly around the vascular cylinder, may be indicative of inherent variation between genotypes in lignin biosynthesis, deposition and/or the chemical composition of lignin. The variation at this location and its relationship to resistance deserve subsequent investigation. In addition, proanthocyanidins does not appear to be a critical part of the defense response in apple root tissues. Our results are consistent with the transcriptome data which suggest that lignin deposition is a major component contributing to resistance of apple root to P. ultimum infection. Further improvement of the sampling strategy and sectioning techniques as well as staining method should provide more consistent comparison between genotypes and infection events. Results from this study are part of the effort to bridge the gap between genomics data, function of specific candidate genes and the whole-plant resistance response. Ultimately, all these data can be used to develop predicative molecular tools for selecting resistant apple rootstock germplasm.
-
The authors confirm contribution to the paper as follows: Zhu Y conceived the study and wrote the manuscript. Rainbow J provided the raw data and involved in writing. Zhou Z participated in the conception of the study and manuscript revision. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
The authors thank India Cain and Southern Reign for initial test of the protocols. This work was supported by USDA ARS base fund.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhu Y, Rainbow J, Zhou Z. 2024. Microscopic features of lignin deposition patterns in young apple roots using brightfield and fluorescence imaging. Fruit Research 4: e007 doi: 10.48130/frures-0023-0045
Microscopic features of lignin deposition patterns in young apple roots using brightfield and fluorescence imaging
- Received: 26 September 2023
- Revised: 04 November 2023
- Accepted: 13 December 2023
- Published online: 01 February 2024
Abstract: Our recent transcriptome analyses have identified that the upregulation of genes involved in phenylpropanoid biosynthesis and laccase-directed lignin formation represent one of the primary defense responses against infection from the necrotrophic oomycete pathogen Pythium ultimum. Taking advantage of lignin autofluorescence in plant tissues and its specific histochemical staining methods, microscopic features of lignin deposition in young apple roots between genotypes and in response to pathogen infection were assessed. The preliminary data indicated that, compared to the Wiesner test, the Maule test is a more efficient lignin staining method for tissue sections of young apple root. Using both brightfield and fluorescence images, elevated intensities of lignin deposition were detected in the parenchyma cells of infected root cortex tissues. The quenched fluorescence intensity due to lignin specific staining corroborated the hypothesis that lignin deposition is a critical component of the detected fluorescence from these sections. The genotype-specific lignin deposition patterns, particularly around vascular bundles, may suggest intrinsic differences in lignin richness and/or monolignol composition between genotypes. Additionally, proanthocyanin deposition was determined to be a less prominent factor in young apple roots. Developing reliable methods to detect anatomical and biochemical changes such as cell wall lignification in response to pathogen infection is crucial for defining the functional role of identified genes and pathways contributing to resistance traits in apple root.
-
Key words:
- Apple root /
- Lignin detection /
- Defense response /
- Resistance trait /
- Microscopic examination /
- Pythium ultimum.












