-
Tea, processed from the leaves of the tea plant [Camellia sinensis (L.) O. Kuntze], is one of the world's most popular non-alcoholic beverages[1]. The appeal of tea stems from the presence of various distinctive secondary metabolites (SMs) such as catechins, theanine, caffeine, volatile chemicals, and so on, all of which have great health benefits and play a significant role in determining the pleasant flavour quality[2]. Thus, understanding how tea produces these special metabolites has become a hot topic. Unlike primary metabolites, SMs are not essential for plant growth and development, while they are related to plant reproduction and stress resistance, and are crucial in managing the interaction between plants and their ecological environment[3]. To date, more than 1,400 specialized metabolites have been isolated and identified in tea[4]. Although most of these are widespread in the plant kingdom with similar functions, in terms of quality and quantity, catechins, caffeine, theanine, and diverse volatile compounds form the unique characteristics of tea[2]. For example, catechins are the source of the bitter and astringent taste of tea; caffeine gives tea the bitter taste; theanine is recognized as an important umami-enhancing compound in tea; and volatile compounds endow tea with abundant and diverse aroma quality[5,6]. Moreover, these distinctive SMs have substantial health benefits, making their metabolism a hot research issue and the primary focus of this paper. As the most abundant ingredients of tea, catechins account for 12%–24% of dry tea[7]. They consist of four major nongalloylated catechins and four galloylated catechins[8]. All of them are derived from the flavonoid pathway[2,9]. Caffeine is the most well-known purine alkaloid and accounts for 2%–5% of dry tea. The key enzymes involved in caffeine biosynthesis [N-methyltransferase (NMT), N-methyl nucleosidase (N-MeNase), theobromine synthase (MXMT), and tea caffeine synthase (TCS)] have been identified[10]. Theanine is a kind of non-protein amino acid, accounting for 1%–2% of dry tea. Recent studies have revealed that alanine decarboxylase (AlaDC) and theanine synthase (TSI) collaborate to regulate the biosynthesis of theanine in tea[11]. Volatile compounds have low content (accounting for less than 0.03% of dry tea) but are fundamental for tea aroma. According to tea volatile biosynthetic pathways, they can be divided into fatty acid derived volatiles (FADVs), volatile terpenes (VTs), and volatile phenylpropanoids/benzenoids (VPBs)[12]. Although the biosynthesis of these SMs follows the specific machinery, significant changes in both number and composition can be detected in varied tea varieties, seasons, geographic locations, or final tea products[13,14]. Many studies also indicated that tea SMs are synthesized to assist tea plants in adapting to their unstable environment and normal growth[15,16]. Taking advantage of this property, many man-made stresses are applied during tea growth or manufacturing processes to 'modify' tea metabolites[6,17,18]. The biosynthesis and accumulation behavior of tea SMs are tightly regulated by their biosynthetic machinery[2], and the changes in target metabolite content arise from variable expression of key synthesis-related genes[19−21]. Therefore, understanding the regulatory mechanisms of gene expression related to SMs biosynthesis is crucial for improving tea quality. With the innovation and development of biotechnology in recent years, great progress has been made in understanding the regulation of secondary metabolism of tea plants at the levels of DNA, mRNA, protein and metabolites[22]. However, regarding the regulation of the transmission of information flow in DNA–RNA–protein, a class of hidden players, non-coding RNAs (ncRNAs), has been ignored for a long time.
The ncRNAs were initially regarded as transcriptional 'noise' as they do not encode proteins or have low protein-coding potential. The latest development of high-throughput sequencing technology and experimental verification shows that ncRNAs are widely involved in the regulation of the expression of protein-encoded genes[23]. The regulatory ncRNAs mainly belong to small RNAs (18–30 nt) and long ncRNAs (> 200 nt)[24]. Specifically, microRNAs (miRNAs) are 21–24 nt in length, constituting a large portion of small RNAs in plants, which function as negative regulators to targets at the post-transcriptional or translation level or mediate DNA methylation[25]. The biogenesis process of miRNAs is widely conserved in plants, including transcription, precursor processing, methylation, and assembly of miRNA-induced silencing complex (miRISC) (as described in detail earlier by Song et al.[26] and He et al.[27]). In addition, long ncRNAs are classed into linear long ncRNAs (commonly known as lncRNAs) and circular long ncRNAs (circRNAs) and are essential modulators of protein-coding gene expression[28]. Each long ncRNA type is synthesized by a specific mechanism and has distinct regulatory properties in cis or trans form (as described in detail earlier by Yu et al.[23] and Xu et al.[29]).
Many studies have found that ncRNA- 'protein-coding gene' interactions play important regulatory roles in plants, such as growth, development, and stress responses[23]. Emerging evidence also suggests that ncRNA-'protein-coding gene' interactions are vital in the modulation of secondary metabolism in horticultural plants[30]. However, there is relatively little research on ncRNAs, especially lncRNAs and circRNAs, concerning the tea plant. We herein summarize recent progress on tea plant miRNAs, lncRNAs, and circRNAs and discuss their regulatory roles in the production of SMs to enlighten the development of novel agronomic tools to improve the quality of tea.
-
miRNAs are the ultimate regulators of gene expression in plants, including tea plants, via direct target mRNA cleavage or translational repression[31,32], which play a key role in the regulation of secondary metabolism[33,34]. In addition, according to the competing endogenous RNA (ceRNA) hypothesis, lncRNAs, circRNAs, and mRNAs can operate as ceRNAs to competitively bind miRNA response elements (MREs) that modulate a variety of life activities[35]. Therefore, miRNAs play a role in the center of secondary metabolic regulatory networks.
The identification of miRNAs in tea plants
-
Twenty-eight articles were retrieved from the Web of Science database (core collection) from 2010 to 2022. The studies on miRNAs in tea can be traced back to 2010. Das & Mondal[36], and Prabu & Mandal[37] identified tea conserved miRNAs and their targets from expressed sequence tags (ESTs) by computational methods. In the same way, Zhu & Luo[38] identified that 14 new miRNAs may target 51 mRNAs in tea, which took part in 13 metabolic networks. In 2012, Mohanpuria & Yadav[39] created a tea small RNA library from a mixture of two leaves and a bud and cloned six novel tea-specific miRNAs. Small RNA sequencing combined with bioinformatics prediction has been employed in several studies to identify novel miRNAs and explore their biological activities since the emergence of second-generation sequencing technology[40−44]. For example, Zhang et al.[40] used Solexa sequencing technology to discover and analyze cold-responsive miRNAs and their targets derived from cold-treated tea leaves. Guo et al.[43] sequenced four small RNA libraries obtained from tea leaves exposed to four different levels of drought treatments. Also, degradome sequencing was frequently used to identify miRNA pairing information with degraded segments of their targets[40−42,45−47]. Due to, however, most miRNAs arising from the intergenic region, studies on the miRNAs identification and functional verification of tea plants progressed slowly and with low precision before the release of the C. sinensis genome. The draft genome of C. sinensis var. assamica 'Yunkang 10' released in 2017 paves the way for more detailed exploration of tea plant miRNAs and their functions[48]. Benefit from the revolution of third-generation sequencing technology, the draft genome of C. sinensis var. sinensis 'Shuchazao' with better assembly continuity was released the following year[49]. The genomes of tea plants were assembled to the chromosomal level[50−54], even to haplotype-resolved levels[55,56], thanks to the availability of high-throughput chromatin conformation capture (Hi-C) technology. This means that raw data yielded from small RNA sequencing can be mapped with high-quality reference genomes, further increasing the accuracy of tea plant miRNA identification and providing a good basis for experimental verification of miRNA functions[57−65]. Because many prediction methods for plant miRNA targets produce a substantial number of false positives in non-modal plants[66], validated miRNA-target interactions can better reflect the actual actions of miRNAs before the potential of miRNAs to regulate SMs biosynthesis can be further considered. As the biogenesis and actions of miRNA have been revealed, many experimental methods such as 5' RLM-RACE, transient co-transformation, northern blot, miRNA-agomir and miRNA-antagomir treatments, etc., have been used to verify miRNA-target interactions[30]. We briefly summarize the currently validated miRNA-target pairs in the tea plant from 18 published articles (Table 1). There were 75 miRNA-target interactions which have been validated at least using the 5'RLM-RACE method. Similar to other plants, most of the miRNAs' targets are TF genes in the tea plant[30], indicating miRNAs have a wide range of regulatory functions. In addition, many miRNA-target pairs specific to tea plants were found to be involved in the regulation of the biosynthesis of SMs. Next, it should be clarified under what conditions miRNAs can regulate the biosynthesis of SMs and we summarize the relevant content below.
Table 1. Summary of validated miRNA-target pairs in tea plant.
miRNA famliy miRNA name Target Action Verification methods Biology functions Reference miR156 miR156f SPL-3 (Squamosa promoter-binding-like protein) Cleavage 5’RLM-RACE; qRT-PCR Colletotrichum gloeosporioides immune response [59] miR156i SBP (S-RNase binding protein) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR156 SBP3 Cleavage 5’RLM-RACE; qRT-PCR; northern blot/co-transformation in tobacco leaves Regulation of the biosynthesis of catechins [62] miR156f-3p SBP; AP2/ERF (Ethylene-
responsive transcription factors)Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR156 SPL Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [67] miR156g-3p F3H (flavanone 3-hydroxylase) Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR159 miR159a GAMYB Cleavage 5’RLM-RACE; qRT-PCR The transformation of plant development stages [34] miR160 miR160a-5p ARF17 (Auxin responsive factor17) Cleavage 5’RLM-RACE; qRT-PCR Abiotic stresses response [45] miR160c ARF Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR160k Dirigent Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR164 miR164a NAC100 (NAC domain transcription factors100) Cleavage 5’RLM-RACE; qRT-PCR Regulation of leaf development [45] miR164a NAC Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR164a NAC-17 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR164a NAC Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR166 miR166 HD-ZIP III (Homeodomain-
leucine zipperIII)Cleavage 5’RLM-RACE; qRT-PCR Drought stress response [69] miR166a HD-ZIP4 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR166d-5p_1 ABCC1-2 (ABC transctipters) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid [61] miR166d-5p_1 ABCG2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [61] miR167 miR167a CHL (Chalcone isomerase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of flavonoid biosynthesis [44] miR167a ARF6 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [44] miR167a ARF8 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [44] miR167 ARF Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [34] miR167d_1 ARF Cleavage 5’RLM-RACE; qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR167d_1 GH3 (Flavanone 3-hydroxylase) Cleavage qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR167d ARF Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR169 miR169e NFY (Nuclear transcription
factor Y)Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR169d-5p_1 ACX (acyl-CoA oxidase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of JA/MeJA and terpenoids [70] miR169a NF-YA (Nuclear
factor Y)Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of theanine and caffeine [62] miR171 miR171b-3p CRE1 (cytokinin receptor 1) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid and terpenoids [61] miR171b-3p_2 DELLA1 (DELLA protein1) Cleavage 5’RLM-RACE; qRT-PCR; miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR172 miR172k ERF_RAP2-7 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR172g-3p MYC2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [61] miR319 miR319c TPC (TCP family transcription factor) Cleavage 5’RLM-RACE C. gloeosporioides immune response [59] miR395 miR395e APS (Aspartic proteases) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR396 miR396b-5p GRF-1 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR396b-5P F3H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of flavonoid [61] miR396 GRF Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [34] miR396d GRF1 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR396d GRF4 Cleavage 5’RLM-RACE; qRT-PCR; northern blot; co-transformation Regulation of the biosynthesis of catechins [62] miR396d GRF13 Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of catechins [62] miR397 miR397 LAC17 (Laccase17) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR398 miR398a-3p-1 CSD4 (Cu/Zn-Superoxide dismutases4) Cleavage 5’RLM-RACE; qRT-PCR Cold stress response [66] miR408 miR408-3p_2ss18GT19GT STK1 (Ser/Thr-protein Kinse) Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR447 miR447g-p5 PAL (Phenylalanine ammonia-lyase) Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] miR529 miR529d CHI (chalcone isomerase) Cleavage 5’RLM-RACE; Regulation of the biosynthesis of catechins [68] miR530 miR530b ERF96 Cleavage 5’RLM-RACE; qRT-PCR; northern blot/co-transformation Infection of Pestalotiopsis-like species response [60] miR530a DHBP Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] miR828 miR828 WER Cleavage 5’RLM-RACE; qRT-PCR Regulation of plant development [45] miR828a MYB (MYB domain protein) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [65] miR828a MYB75 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR845 miR845 ABCC1-3 Cleavage 5’RLM-RACE; qRT-PCR/miRNA-agomir/antagomir Regulation of the biosynthesis of flavonoid and terpenoids [61] miR845 ABCC2 Cleavage qRT-PCR; miRNA-agomir/antagomir Inhibite flavonoid biosynthesis but enhance terpenoid biosynthesis [61] miR858 miR858a MYB12 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of polyphenols [45] miR858b_R-3 MYB Cleavage 5’RLM-RACE; qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR858b MYB Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of terpenoids [65] miR858a R2R3-MYB Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [59] 858b_R-3 MYB114 Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides immune response [47] miR2593 miR2593e ANR (Anthocyanidinreductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [44] miR2868 miR2868 LAR (leucoanthocyanidin reductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR3444 miR3444b DFR (Dihydroflavonol 4-reductase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [44] miR3630 miR3630-3p_L-3p MYC Cleavage qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] miR4380 miR4380a DFR Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR5240 miR5240 DFR Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR5251 miR5251 C4H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR5559 miR5559-5p ANR1; ANR2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR5564 miR5564 ANR2 Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [68] miR7777 miR7777-5p C4H Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of catechins [62] miR7814 miR7814 CHS1 (chalcone synthase1) Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR7814 CHS3 Cleavage 5’RLM-RACE Regulation of the biosynthesis of catechins [68] miR8757 miR8757f PPO (Polyphenol oxidase) Cleavage 5’RLM-RACE; qRT-PCR Infection of Pestalotiopsis-like species response [60] novel miRNA miRn211 TLP (thaumatin-like protein) Cleavage 5’RLM-RACE; qRT-PCR; northern blot; co-transformation Infection of Pestalotiopsis-like species response [60] PC-5p-80764_22 WRKY (WRKY transcription
factor)Cleavage 5’RLM-RACE; qRT-PCR C. gloeosporioides and abiotic stresses response [47] PC-3p-81-33418 AP2/ERF Cleavage qRT-PCR; northern blot Regulation of the biosynthesis of terpenoids [65] novel_miR44 LOX (Lipoxygenase) Cleavage 5’RLM-RACE; qRT-PCR Regulation of the biosynthesis of JA/MeJA and terpenoids [70] miRNA-target pairs modulate the biosynthesis of SMs in the tea plant
-
miRNAs can negatively modulate the content of related SMs in plants by directly regulating the expression of target genes in SM biosynthesis pathways. However, the opposite may be true if the two metabolites belong to different synthetic branches of a metabolic pathway and have a common synthetic precursor[39]. For instance, Sharma et al.[71] identified that miR858 can positively modulate the content of flavonoids, because inhibiting the expression of miR858 can increase the expression level of its targets MYBs, which results in the metabolic flow redirecting to the flavonoid synthesis at the expense of lignin synthesis. MiRNAs can regulate the amount of a class of chemicals belonging to the same metabolic pathway because the majority of their target genes are TFs[33]. While the roles of structural genes, in general, is not as extensive as that of TFs, miRNA-structural gene interactions are more targeted for regulating specific SM content. For instance, the nta-miRX27-QPT2 interaction specific to Nicotiana tabacum has been confirmed to have a regulatory function in nicotine accumulation[72]. In tea plants, miRNA-target pairs modulate the biosynthesis of SMs show lineage-specific target genes.
Modulation of the biosynthesis of catechins
-
An illustration of the general flavonoid pathway leading to the biosynthesis of catechins has been described (Fig. 1). Their accumulation is affected by developmental cues and environmental stimuli[73]. During which miRNAs are confirmed to be involved in the modulation of catechins (Fig. 1).
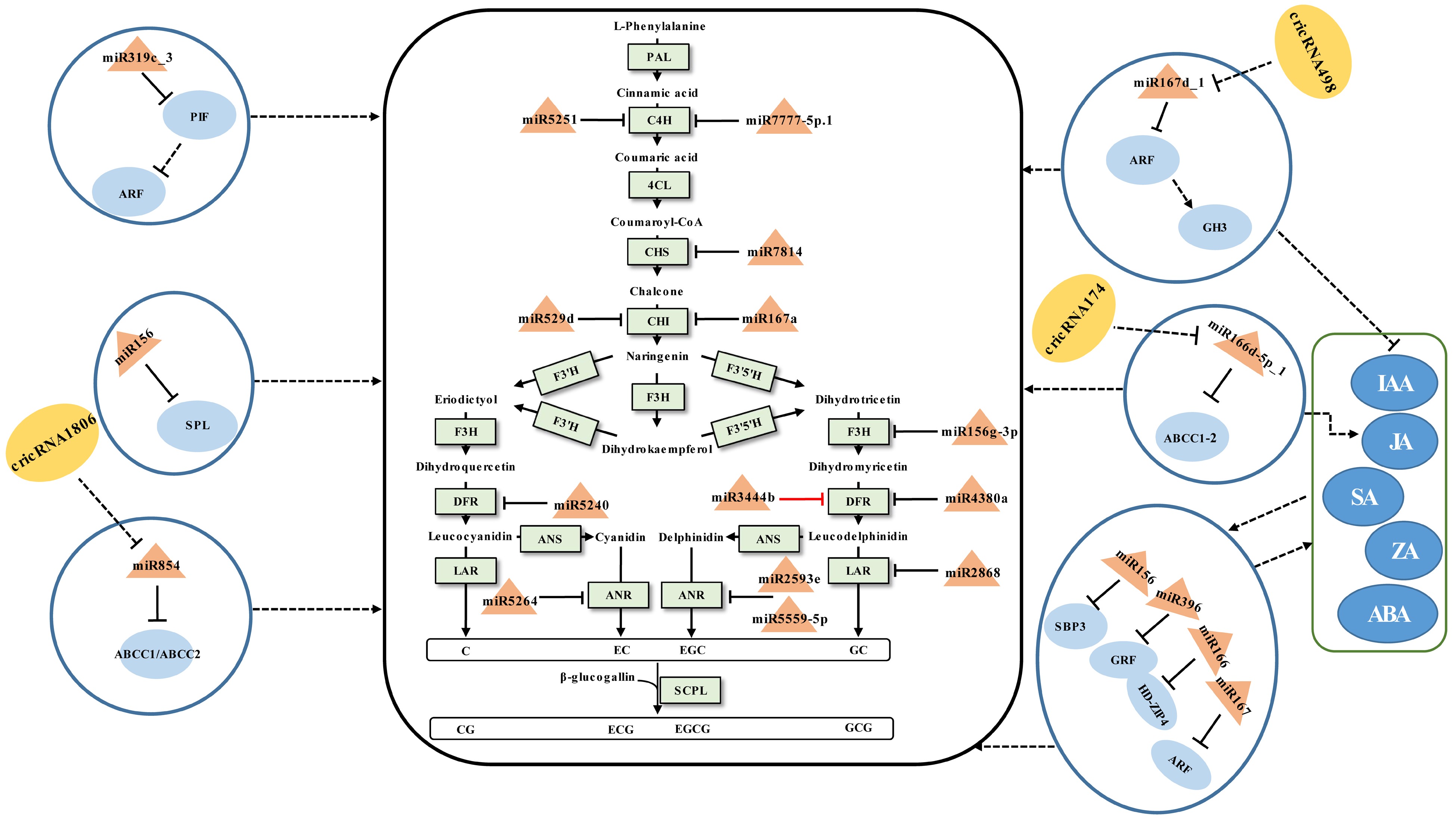
Figure 1.
Schematic representation of the biosynthesis of catechins regulated by ncRNAs in tea plants. The miRNA, circRNA, transcription factor gene, structural gene, and phytohormone are marked in the orange triangles, yellow ovals, light blue ovals, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; the black and red T-shaped lines represent negative and positive regulation, respectively; the black dashed T-shaped line is supported by less experimental evidence or prediction results. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
The content of catechins varies between tissues of the tea plant, which can be mediated by miRNAs. In general, the total amount of catechins collected in young leaves and buds was greater than that in mature leaves[34,44,62]. Sun et al.[44] investigated the functions of miRNAs in modulating the accumulation of catechins in tea buds, first to fifth leaves, and mature leaves. It was found that miRNA167a-CHI, miR2593e-ANR, miR4380a-DFR, miR3444b-DFR, csn-miR5251-C4H, and miR7777-5p.1-C4H pairs may be involved in the regulation of catechin accumulation in tea leaves of different maturity by integrated 5'RLM-RACE and qRT-PCR analyses[44]. Among these, miR3444b and DFR showed a similar expression trend instead of an opposite expression pattern. Considering the mode of action of miRNAs negatively regulating targets, the effect of miR3444b-DFR on the regulation of catechins needs to be further validated. At the same time, miR529d and miR156g-3p were shown to be involved in the regulation of catechin accumulation in the first, third, and oldest leaves of the tea plant, respectively, via targeting CHI and flavanone 3-hydroxylase (F3H)[68]. Although the miR7814-CHS1, miR7814-CHS3, miR5240-DFR, miR2868-LAR, miR5559-5p-ANR1, miR5559-5p-ANR2, and miR5264-ANR2 interactions that exist in tea plants have also been validated, they were not related to the regulation of differential accumulation of catechins in different leaf positions[68]. This requires further elucidation of the regulatory role of these miRNAs on catechins synthesis under what conditions, such as abiotic stress and biotic stress. In addition to structural genes, miRNA-TF gene modules have been implicated in the control of catechin accumulation. Zhao et al.[62] conducted a thorough investigation to explore the regulatory mechanisms that miRNA-mediated the biosynthesis of taste compounds in the different tea leaf positions. It was found that miR156, miR164a, miR166a, miR167d, and miR396d may negatively regulate the the accumulation of catechins by targeting S-RNase binding protein3 (SBP3), NAC domain transcription factor (NAC), homeodomain-leucine zipper4 (HD-ZIP4), auxin responsive factor (ARF), and growth-regulating factor1, 4, and 13 (GRF1, 4, and 13) cleavage, respectively. These modules and phytohormones, such as indole-3-acetic acid (IAA), jasmonic acid (JA), abscisic acid (ABA), zeatin (ZA), and salicylic acid (SA), might synergistically regulate the biosynthesis of catechins although there is no direct evidence to support this claim[62]. Li et al.[34] studied the regulatory link between essential taste components and miRNAs in different tissues of the tea plant. According to integrated 5'RLM-RACE and qRT-PCR validation results, three miRNA-target pairs (miR159-GAMYB, miR167-ARF, and miR396-GRF) may cause the distinct accumulation of catechins, yet this has not been verified by experiments.
The accumulation of catechins in tea leaves differs depending on the nitrogen form, which can be regulated by miRNAs. miR156 regulated the accumulation of catechins in tea shoots by inhibiting the expression of the target SPL, a TF gene that promote the expression of DFR[67]. In the presence of
$\text{NO}^-_3 $ $ \text{NH}^+_4$ miRNAs can mediate the biosynthetic modulation of catechins during the manufacturing processes of postharvest tea leaves. The accumulation of SM in tea leaves during the preharvest stage serves as the foundation for developing finished tea quality, while the manufacturing processes of postharvest tea leaves determine the final flavor quality of the finished tea. Especially in withering and turning-over processes, tea leaves remain alive under multi-stresses[74]. During this time, miRNAs have been discovered to control catechin metabolism via miRNA-TF gene modules. Solar-withering is a crucial process in tea production that can improve the palatability of tea by reducing catechin content appropriately[70]. Tea leaves are subjected to UV irradiation, heat, and dryness during the withering process, which can cause stress responses that are mediated by miRNAs to have an impact on the accumulation of related SMs[61]. The accumulation of catechins was linked to the miR845-ABCC1-3/ABCC2, miR319c_3-PIF-ARF, miR166d-5p_1-ABCC1-2, and, miR167d_1-ARF-GH3 modules[61]. These suggested that when the tea plant is under abiotic stress, miRNAs may indirectly regulate the accumulation of catechins by regulating genes related to phytohormone synthesis or metabolite transport.
Modulation of the biosynthesis of caffeine and theanine
-
Caffeine biosynthesis in tea plants is relatively clear[2] (Fig. 2). The regulatory mechanism of protein-coding genes affecting caffeine synthesis has been widely explored[20,75], but there are relatively few studies on the regulatory mechanism of miRNA affecting caffeine synthesis. Similarly, the study of theanine metabolism in tea plants has achieved a breakthrough[11,76] (Fig. 2), but the regulatory miRNAs involved in theanine biosynthesis remain largely unknown. To date, only one case of validated miRNA-target pair being involved in regulating caffeine and theanine synthesis has been reported[62]. The miR169 was proved to target NF-YA cleavage by 5'RLM-RACE. The decrease of miR169 expression from the tea bud to the fourth leaf will enhance the expression of its target NF-YA, indicating that the expression level of miR169 positively linked with the content of caffeine and theanine. According to weighted gene co-expression network analysis (WGCNA), it was found that both the content of IAA, JA, ZA, and ABA and the content of caffeine and theanine had significant correlations with the expression of miR169. It suggested that miRNA-mediated modulation of phytohormone crosstalk during specific developmental processes may modulate the caffeine and theanine biosynthesis. Of course, interesting findings require more experiments for verification.
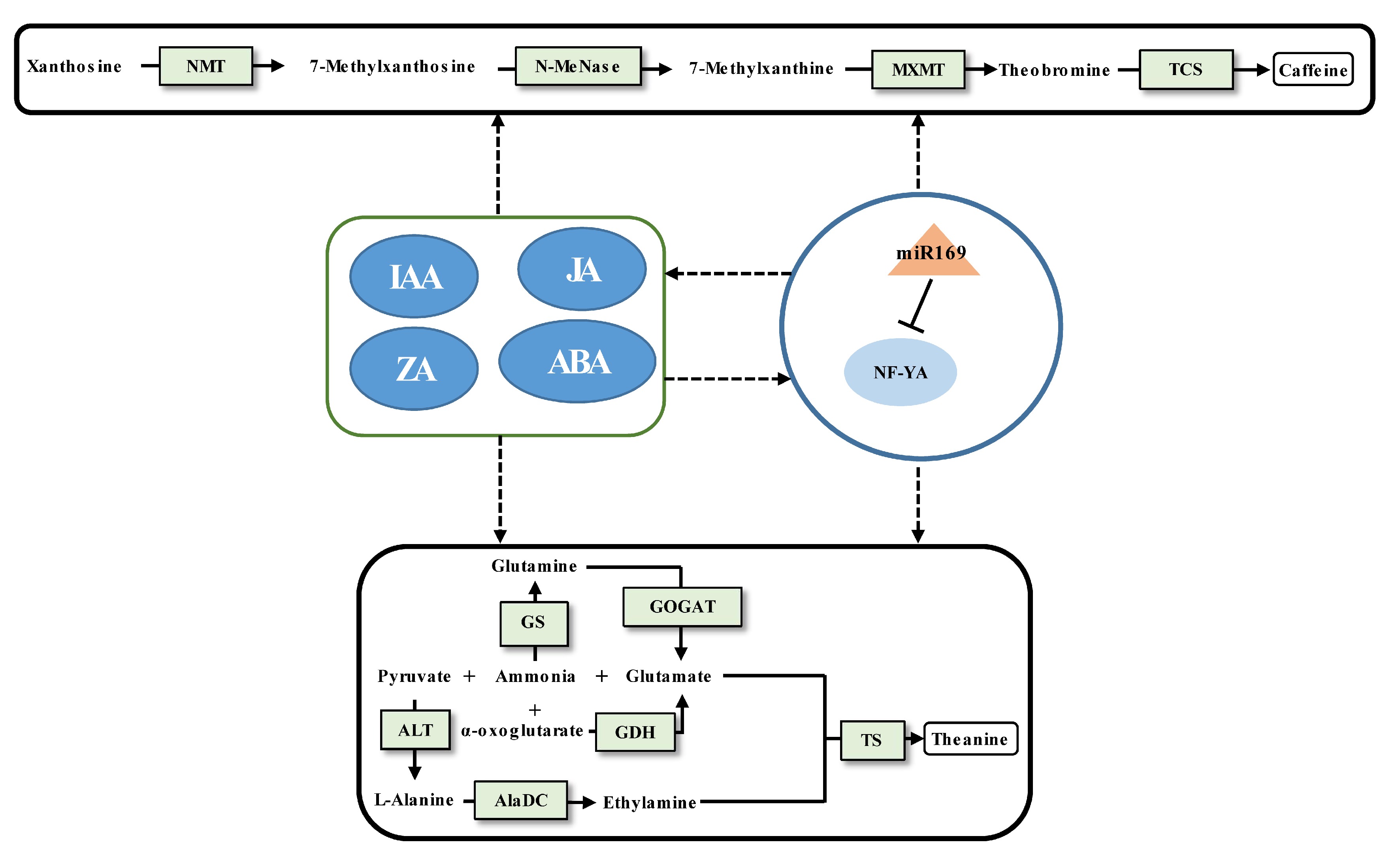
Figure 2.
Schematic representation of the biosynthesis of caffeine and theanine regulated by ncRNAs in tea plants. The miRNA, transcription factor gene, structural gene, and phytohormone are marked by the orange triangle, light blue oval, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; and the black T-shaped line represents negative regulation. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
Modulation of the biosynthesis of VTs
-
VTs mainly include monoterpenes and sesquiterpenes, which can be synthesized via the plasmatic methylerythritol phosphate (MEP) pathway and the cytosolic mevalonic acid (MVA) pathway (Fig. 3). They are the primary components of floral and fruity aromas in tea[12]. The content of VTs in preharvest tea leaves are relatively low. Both the environmental stimuli and the abiotic stresses induced by the postharvest manufacturing processes can lead to tea leaves accumulating high levels of VTs. It has recently been shown that miRNAs regulate to the biosynthesis of VTs in tea leaves.
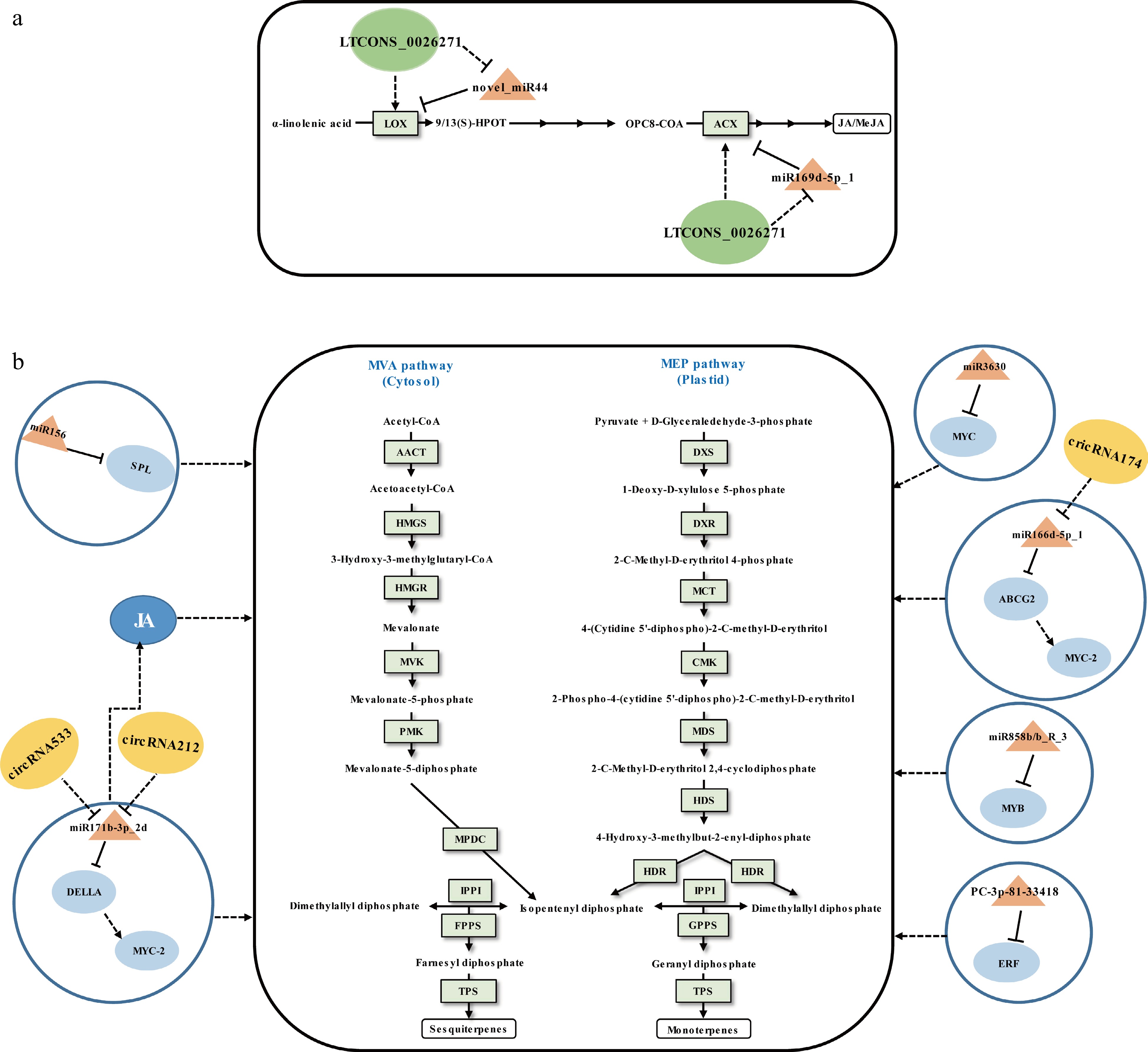
Figure 3.
Schematic representation of the biosynthesis of (a) JA/MeJA and (b) volatile terpenoids in tea plants. The miRNA, circRNA, lncRNA, transcription factor gene, structural gene, and phytohormone are marked in the orange triangles, yellow ovals, green ovals, light blue ovals, light green boxes, and dark blue ovals, respectively. The solid line with the arrow indicates direct regulation; the dashed line with the arrow is supported by less experimental evidence or prediction results; the black T-shaped line indicates negative regulation; the black dashed T-shaped line is supported by less experimental evidence or prediction results. The miRNA-target pairs shown in the figure have been verified to have regulatory relationships.
Tea plant miRNAs can mediate the regulation of differential synthesis of VTs in different growing stages. Zhao et al.[65] investigated the relationship between miRNAs and VTs and found that four validated miRNA-target interactions, including miR156-SPL, miR3630-MYC, miR858b_R-3-MYB, and PC-3p-81_33418-ERF, may be involved in VTs accumulation at different growing time points. However, due to the lack of experimental evidence of direct interaction between TFs and structural genes related to VT synthesis, the above results are only speculative based on WGCNA.
Tea plant miRNAs can intervene in the biosynthetic regulation of VTs during withering. Zhu et al.[70] validated that novel_miR44 and miR169d-5p_1 can negatively regulate JA biosynthesis by cleavage of the upstream structural genes of JA synthesis LOX (lipoxygenase) and ACX (acyl-CoA oxidase), respectively (Fig. 3a)[70]. Many studies have shown that JA promotes the biosynthesis of VTs in plants[77], which may be crucial for increasing the content of VTs. Moreover, miR166d-5p_1-ABCG2-MYC2 and miR171b-3p_2-DELLA-MYC2 modules were involved in VTs biosynthesis indirectly[61] since the MYC2 acts as a major regulator of JA biosynthesis[78]. Due to the fact that miRNAs have a wide range of regulatory functions when their targets are TFs, both miRNAs (miR166d-5p_1-ABCG2-MYC2 and miR171b-3p_2-DELLA-MYC2 modules), acting as positive regulators, have boosted VT accumulation in sunlight-withered leaves (Fig. 3b). Undoubtedly, the indirect regulatory functions of miRNAs on SMs need further research for verification.
-
Tea leaves are subjected to several abiotic stresses during postharvest processing. Because the leaf cells are alive, transcriptional alterations would have a significant impact on the accumulation of SMs[79]. Many plants have been reported to have lncRNAs and circRNAs that are involved in the abiotic stress response[80]. The large potential of lncRNAs and circRNAs in the modulation of SMs biosynthesis is also gradually being taken seriously by tea scientists.
lncRNAs that participate in the biosynthesis of SMs in tea plants
-
Many studies have shown that lncRNAs play important roles in SMs biosynthesis by regulating related gene expression in cis or trans[72,81,82]. Because a large part of lncRNAs arises from non-coding regions, there was no research on tea plant lncRNAs until 2019, i.e. after the publication of the tea plant genome sequences[83]. So far, to the best of our knowledge, there have been eight articles published concerning tea plant lncRNAs[70,83−89], which showed that lncRNAs were involved in response to pathogenic microorganisms attack[85,87−89], salt stress[84], and regulation of secondary metabolism[70,83,86]. We focus on lncRNAs that relate in this section. Varshney et al.[83] first employed the bioinformatics prediction method that identified 33,400 lncRNAs of tea plants based on 170 public RNA-seq data from 11 harvestable tissues. That same year, Zhu et al.[70] performed a systemic analysis of lncRNAs during tea production (Fig. 3b). It was identified that 32,036 lncRNAs from fresh leaves, solar-withered leaves, and indoor-withered leaves of tea plant. An analysis of the differentially expressed lncRNAs and their target genes were identified as related to flavonoid metabolism as well as terpenoid accumulation. In addition, by integrated bioinformatics prediction and qRT-PCR analysis, lncRNAs LTCONS_00026271 and LTCONS_00020084 were preliminarily proven that can serve as the endogenous target mimics (eTMs) of novel_miR44 and miR169d-5p_1, respectively, that promote the biosynthesis of VTs in solar-withered leaves (Fig. 3). In another study, a total of 1,679 lncRNAs were discovered from the full-length transcriptome sequencing data of shoot tips and roots of tea plants. The bioinformatics prediction showed that lincRNAs are closely related to the regulation of characteristic secondary metabolites, such as catechins, theanine, and caffeine by influencing the expression of related genes[86]. These results suggested that lncRNAs are ubiquitous regulators in tea plant SMs biosynthesis.
circRNAs involved in the biosynthesis of SMs in tea plants
-
The potential involvement of circRNAs in the modulation of biosynthesis of SMs in plants has been explored in Salvia miltiorrhiza[90]. In 2018, Tong et al.[91] first pursued an analysis of circRNAs in the tea plant and identified 342 high-confidence circRNAs. The expression levels of circRNAs were positively correlated with the expression levels of their parental transcripts in different mature leaves. Moreover, the gene sets encoding circRNAs revealed their candidate roles in metabolite biosynthesis base on the KEGG enrichment analysis. Subsequently, the circRNAs functioning as miRNA sponges during the tea withering process was predicted in another study. Combined with qRT-PCR analysis, circRNA498, circRNA1806, circRNA212/circRNA533, and circRNA174 were preliminarily proved that can mediate flavor-related metabolites by sponging corresponding miRNAs related to phytohormone signaling and ATP binding cassette (ABC) transporters[61] (Fig. 1 & Fig. 3b). Given the limited number of studies on tea circRNAs, the above-described circRNA functions may not reflect their general rules in the regulation of SMs biosynthesis in tea. In the future, functional studies will be useful for uncovering their regulatory functions.
-
We summarized recent research progress in the biosynthetic regulation of tea SMs exerted by ncRNAs. In general, the mechanisms by which tea plant miRNAs regulate the biosynthesis of SMs have been demonstrated by some experimental evidence. It is also encouraging to note that the potential of lncRNAs and circRNAs in regulating SM biosynthesis in tea have also received increasing attention from scholars. The rapid development of second-generation sequencing technology and the increasingly powerful bioinformatics tools accelerating the application of large-scale sequencing efforts to functional studies of tea plant ncRNAs. As data accumulates, it will be exciting to witness further developments in our understanding of the functional roles of ncRNAs in the biosynthesis modulation of SMs in tea. Future research on tea plant ncRNAs can be conducted mainly on the following aspects:
(1) Establishment of a tea plant ncRNA database is needed. To facilitate the use and mining of plant ncRNA data, many published ncRNA databases have been established. However, the tea plant has also evolved a variety of specific ncRNA-mediated regulatory mechanisms on unique SMs[44,68]. To date, it is inconvenient to obtain detailed information such as the sequences, chromosome locations, functions, and annotations when researchers look at particular ncRNA-target interactions involved in the biosynthesis modulation of SMs in tea plants. Hence, a public database for tea plant ncRNA sequences and annotation are urgently required.
(2) More experimental methods need to be applied to validate the functions of ncRNAs-mediated biosynthetic regulation of SMs in tea. Generally, plant miRNAs are classified into conserved miRNAs, less conserved miRNAs, and species-specific miRNAs that are present in angiosperms, a lineage or group of plants, and a single species, respectively[92]. While lncRNAs have an even lower degree of conservation among plant kingdoms[93]. To date, the specific functions of many ncRNAs have been clarified in model plants such as A. thaliana, Oryza sativa, N. tabacum, etc., which have stable genetic transformation systems[23,26]. Compared with ncRNA studies in model plants, progress in tea plants is relatively slow, especially because of the lack of a tea plant genetic transformation system for functional verification of ncRNAs. As described above, most regulatory mechanisms of ncRNA-mediated accumulation of tea SMs were indirectly demonstrated by qRT-PCR combined with bioinformatics analysis, or other in vitro experiments[61,70]. Several in vivo functional identification of miRNAs have used heterologous expression systems or tea transient transformation systems[57,60,61]. To make ncRNAs promising regulatory tools for tea quality improvement, the specific regulatory roles of tea plant ncRNAs on secondary metabolism in vivo need to be analyzed in detail. Recently, an efficient mesophyll protoplast isolation method for tea has been established[94], which provided a foundation for chitosan-complexed carbon nanotube carriers and nanoparticles with polyethylene glycol-mediated protoplast transformation in tea plants in the future[95]. In addition, an in planta transformation approach for gene functional verification of tea plants has been reported recently[96]. In the future, it is believed that vector construction and genetic transformation based on the in planta transformation system opens an intriguing research area to verify the specific ncRNA-target functions in tea plants.
(3) The miRNAs have the potential as important agronomic tools to improve tea quality. If the expected quality of tea is to be improved via conventional breeding methods, long-term efforts are needed to identify and introduce beneficial traits into the tea plants. Moreover, the lack of an effective genetic transformation system for tea plants makes the advantage of transgenics over traditional breeding in terms of time disappear. Recently, it has been found that the first ORF after the transcription start site of plant primary miRNA (pri-miRNAs) corresponded to a translated ORF coding regulatory peptides called miRNA-encoded peptides (miPEPs), able to increase the expression of pri-miRNAs[97]. Ormancey et al. proved that watering plants with miPEPs can lead to an overall change in plant development by elevating the expression of corresponding miRNAs and, correlatively, decreasing the expression of the respective miRNA target genes, thus being a suitable alternative to the use of chemicals in agronomy[98]. While we are working on the molecular mechanisms of tea quality regulation, are there any methods that have the potential to be quickly and cost-effectively applied to the quality improvement of tea? Let's make a hypothesis. Catechins undoubtedly have a wide range of health benefits for the human body[99]. In addition, catechins are responsible for bitter and astringent tastes in the tea infusion[8]. Thus, moderately reducing the content of catechins may improve the palatability of tea. Many miRNAs have been found to negatively regulate the accumulation of catechins[44,68]. Suppose that their nascent pri-miRNAs have the potential to encode miPEPs. In that case, we can imagine using a cocktail of several miPEPs to increase the expression of their nascent pri-miRNAs and decrease the expression of the respective miRNA targets correlatively, thereby achieving the fine-tuning of flavor quality of tea.
This work was supported by the 6.18 Tea Industry Technology Branch of Collaborative Innovation Institute (K1520001A), the Construction Project for Technological Innovation and Service System of Tea Industry Chain of Fujian Agriculture and Forestry University (K1520005A01), the Rural Revitalization Tea Industry Technical Service Project of Fujian Agriculture and Forestry University (11899170145), the 'Double First-class' Scientific and Technological Innovation Capacity and Enhancement Cultivation Plan of Fujian Agriculture and Forestry University (KSYLP004), the Construction of Plateau Discipline of Fujian Province (102/71201801101), and the Tea Industry Branch of Collaborative Innovation Institute of Fujian Agriculture and Forestry University (K1521015A). The authors thank Xin Huang, Master of Translation, from the School of Foreign Languages of Fuzhou University for her linguistic assistance.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhou C, Tian C, Zhu C, Lai Z, Lin Y, et al. 2022. Hidden players in the regulation of secondary metabolism in tea plant: focus on non-coding RNAs. Beverage Plant Research 2:19 doi: 10.48130/BPR-2022-0019
Hidden players in the regulation of secondary metabolism in tea plant: focus on non-coding RNAs
- Received: 13 July 2022
- Accepted: 28 September 2022
- Published online: 02 November 2022
Abstract: Non-coding RNAs (ncRNAs) are functional transcripts with minimal or no protein-coding capacity that comprise a large portion of the plant transcriptome. Among them, the microRNAs (miRNAs), linear long ncRNAs (lncRNAs), and circular long ncRNAs (circRNAs) have been widely proven to play essential regulatory roles in the biosynthesis of secondary metabolites (SMs) by modulating the expression of key synthesis-related genes in plants. Tea boasts numerous characteristic SMs, such as catechins, theanine, caffeine, volatile compounds, etc., which have distinguished health properties and largely determine the pleasant flavor quality. Thus, understanding how the tea plant produces these specialized metabolites is of great research interest. With the innovation and progress of biotechnologies in recent years, significant progress has been made in research on the regulation mechanism of SMs in tea plants at the DNA, mRNA, protein and metabolite levels. The release of the genome sequences of tea plants paves a path for precisely exploring ncRNAs and their functions in tea, and their huge potential for the biosynthesis regulation of SMs has gradually received attention. We herein summarize recent progress on miRNAs, lncRNAs, and circRNAs in tea plants and discuss their regulatory roles in the accumulation of SMs to enlighten the development of novel agronomic tools to enhance the quality of tea.
-
Key words:
- Camellia sinensis /
- miRNA /
- Secondary metabolite /
- Non-coding RNA













