-
Seed size is an important agronomic trait for domesticating crops. Increasing seed size is an ongoing target for improving yield. Seed development in angiosperms is launched by double fertilization within the mature ovule, which leads to the development of a diploid embryo and a triploid endosperm. The seed coat is a maternal sporophytic tissue originating from the ovule integuments. Therefore, seed size is coordinately regulated by the growth and development of the embryo, endosperm, and seed coat.
Seed coat and endosperm growth precede embryo growth during early seed development[1,2]. The seed coat not only delivers nutrients to the endosperm and embryo but also acts as the physical constraint on endosperm and embryo growth[2−4]. The growth of the endosperm, in turn, promotes elongation of the seed coat cell[4,5]. It has been proposed that the seed coat and the endosperm act coordinately to set the volume of the seed cavity for later embryonic growth and determining the seed size in Arabidopsis[2,5]. In rice and many other monocot plants, grain size is also influenced by growth of the maternal tissue spikelet hull and the endosperm[6]. Thus, the maternal tissues and endosperm play primary roles in controlling seed size. Several recent studies have demonstrated that the initiation and correct development of the seed coat depend on the endosperm rather than the embryo, and that early endosperm development is an autonomously programmed process independent of embryogenesis in Arabidopsis[7,8]. The fertilized endosperm of the embryo-free seed in Arabidopsis undergoes normal syncytium formation and cellularization as that of the wild type in terms of the cytological process and time course[7]. Additionally, an increase in coenocytic endosperm turgor pressure drives expansion of the seed[9,10]. In this case, the endosperm plays a central role in seed development and determining seed size.
Development of the nuclear endosperm is a common mechanism among angiosperms, including the monocot cereals and most dicot plants, and is characterized by the rapid proliferation of endosperm nuclei without cell division leading to the generation of a large syncytium during early endosperm development (Fig. 1a, b). Cellularization of the syncytial endosperm is initiated in the micropylar endosperm of Arabidopsis after the globular embryo stage[11]. After cellularization, endosperm cells undergo a small number of synchronous cell division depending on their position along the micropylar-chalazal axis[12]. The central portion of the peripheral endosperm undergoes cell division until the central cell cavity is completely filled with cells[12]. The endosperm then begins to break down gradually, and the reserves support early embryo development[13]. Finally, only a single peripheral endosperm cell layer, referred to as the aleurone layer, is present in the mature seed[13] (Fig. 1a). The rate of division of the cellularized endosperm is much slower than that of syncytial endosperm nuclei[1,14]. The seed almost reaches the final size at the late globular stage of embryo development[2]. Coincidence in the timing of endosperm cellularization with the end of the main stage of seed growth indicates that proliferation of the syncytial endosperm and the timing of endosperm cellularization play crucial roles in determining the sizes of the endosperm and seed in Arabidopsis and many other dicot plants[12] (Fig. 1a). The endosperm of monocot plants, such as maize and rice, occupies the most volume in the mature seed (Fig. 1b). After growth of the ephemeral syncytial endosperm, the endosperm undergoes rapid cellularization and differentiation. During this time, the cellularized endosperm also undergoes a small series of cell proliferation. Then, endoreduplication and cell expansion occur in the central part of the endosperm. Finally, the endosperm cells undergo programmed cell death (PCD) and desiccation (Fig. 1b). During the first 10 days after pollination (DAP), the proliferation of syncytial endosperm nuclei and the endosperm cell division to form the majority of the endosperm cells, which determines kernel sink strength, storage capacity, and kernel size[1,15,16] (Fig. 1b). Therefore, early endosperm growth plays a fundamental role in determining seed size in higher plants. Here, we focus on the recent advances in the regulation of early endosperm growth and discuss the possible molecular mechanisms by which early endosperm development controls seed size (Fig. 2; Table 1).
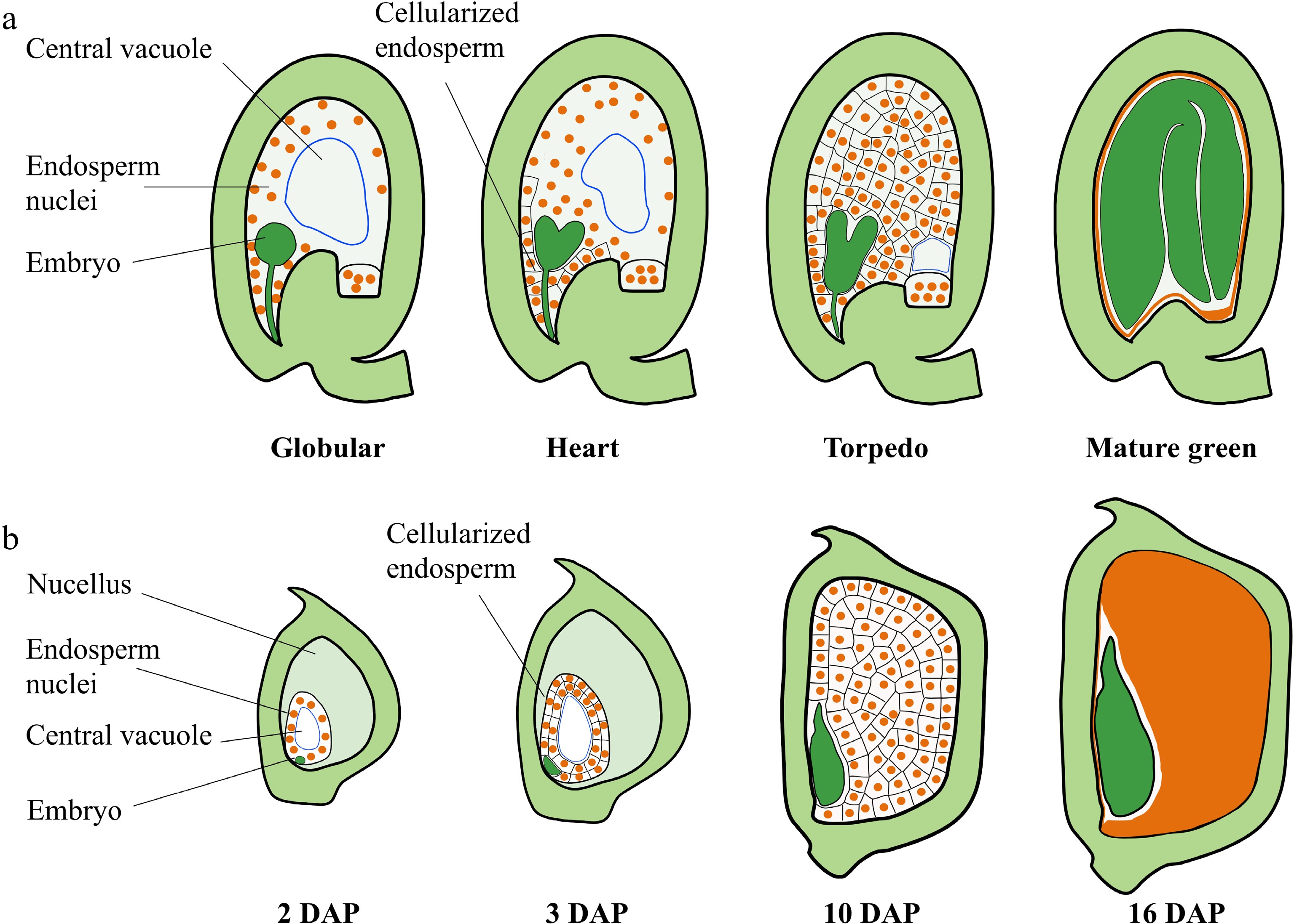
Figure 1.
Seed development in Arabidopsis and maize. (a) Schematic representation of seed development in Arabidopsis. Stages of embryo development are indicated. After fertilization, the endosperm nucleus undergoes rapid division without formation of cell walls or cytokinesis, resulting in a syncytium. Cellularization is initiated in the micropylar endosperm at the early heart embryo stage and is completed when the embryo reaches the torpedo stage. The cellularized endosperm undergoes a small series of cell proliferation until the central cell cavity is completely filled with cells. Then, the endosperm begins to breakdown, and the seed cavity is replaced by the embryo. (b) Schematic representation of seed development in B73 maize. Stages indicate days after pollination (DAP). After the syncytial endosperm proliferation and cellularization, the cellularized endosperm undergoes endoreduplication (starting at 8–10 DAP), followed by PCD (starting at about 16 DAP). The endosperm occupies the largest part of the mature kernel.
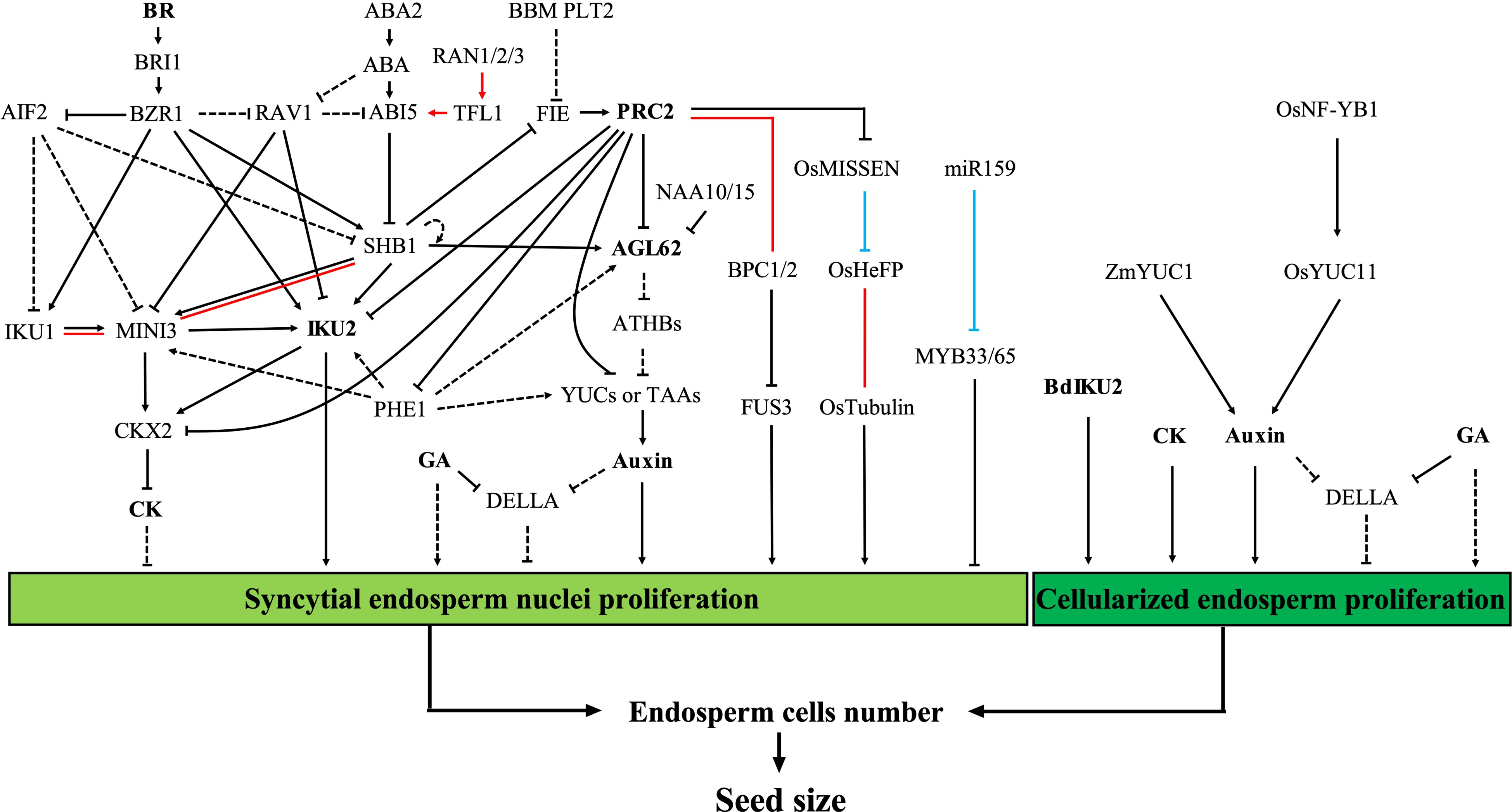
Figure 2.
Regulation of seed size by early endosperm development. Proliferation of the syncytial endosperm nuclei and the endosperm cells during early endosperm development determines the number of endosperm cells, which plays a fundamental role in the control of seed size. The red lines represent protein-protein interactions. The blue lines represent post-transcriptional regulation. The dashed lines represent unconfirmed relationships in the endosperm or in the Arabidopsis endosperm (relationships between 'AGL62', 'ATHBs', and 'YUCs or TAAs'). Abbreviations: ABA, abscisic acid; BR, brassinolide; CK, cytokinin; GA, gibberellin; PRC2, polycomb repressive complex 2.
Table 1. Genes involved in seed size control by early endosperm development.
Species Gene name Accession number Gene product Reference(s) Arabidopsis ABA2 AT1G52340 Short-chain dehydrogenase/reductase; involved in ABA biosynthesis [35] Arabidopsis ABI5 AT2G36270 bZIP transcription factor [34, 35] Arabidopsis AGL62 AT5G60440 MADS-box transcription factor [22, 23, 33,
40, 43, 44]Strawberry FveAGL62 FvH4_2g03030 MADS-box transcription factor [44] FveAGL80 FvH4_6g08460 Arabidopsis AHK2 AT5G35750 Histidine kinase; cytokinin receptor [62] AHK3 AT1G27320 AHK4 AT2G01830 Arabidopsis AHP2 AT3G29350 Histidine phosphotransfer protein; regulator of cytokinin signaling [63] AHP3 AT5G39340 AHP5 AT1G03430 Arabidopsis AIF2 AT3G06590 Non-DNA-binding bHLH transcription factor [70] Arabidopsis ARR1 AT3G16857 Transcription factor; involved in cytokinin signaling [64] ARR10 AT4G31920 ARR12 AT2G25180 Strawberry FveATHB29b FvH4_5g17830 ATHB subfamily of transcription factor [44] FveATHB30 FvH4_6g48610 Arabidopsis AXL AT2G32410 Subunit of the RUB1 activating enzyme; involved in auxin signaling [38, 39] AXR1 AT1G05180 Arabidopsis BBM AT5G17430 AP2/ERF transcription factor [25] Arabidopsis BPC1 AT2G01930 BPC transcription factor [100] BPC2 AT1G14685 Arabidopsis BZR1 AT1G75080 Transcription factor; involved in BR signaling [69] Arabidopsis CKI1 AT2G47430 Histidine kinase without cytokinin perception domain; involved in cytokinin signaling [61] Arabidopsis CKX2 AT2G19500 Cytokinin oxidase; involved in cytokinin homeostasis [27] Rice OsEMF2a Os04g08034 Polycomb group protein [53, 95] Arabidopsis FIE AT3G20740 Polycomb group protein [22, 24] Rice OsFIE1 Os08g04290 Polycomb group protein [94] Rice OsFIE2 Os08g04270 Polycomb group protein [90, 91, 93] Arabidopsis FIS2 AT2G35670 Polycomb group protein [24, 40, 43] Arabidopsis FUS3 AT3G26790 B3 domain transcription factor [100] Arabidopsis IKU1 AT2G35230 VQ motif protein [17−19, 27] Arabidopsis IKU2 AT3G19700 LRR-RLK [17, 19−21, 26, 27] Maize ZmIPT2 Zm00001d003869 Isopentenyl transferase; involved in cytokinin biosynthesis [57, 60] Arabidopsis MEA AT1G02580 Polycomb group protein [24, 43] Soybean GmMEA Gm11G067000 Polycomb group protein [26] Arabidopsis MINI3 AT1G55600 WRKY transcription factor [18−21, 27] Arabidopsis MIR159A AT1G73687 microRNA159 [31] MIR159B AT1G18075 MIR159C AT2G46255 Maize Zma-miR169o MI0013202 microRNA169o [15] Arabidopsis MSI1 AT5G58230 Polycomb group protein [24] Rice MISSEN XLOC_057324 Long noncoding RNA [32] Arabidopsis MYB33 AT5G06100 MYB transcription factor [31] MYB65 AT3G11440 Arabidopsis NAA10 AT5G13780 Catalytic subunit of Arabidopsis NatA complex [33] Arabidopsis NAA15 AT1G80410 Auxiliary subunit of Arabidopsis NatA complex [33] Maize ZmNF-YA13 Zm00001d018255 Nuclear factor Y, subunit A [15] Rice OsNF-YB1 Os02g49410 Nuclear factor Y, subunit B [46] Arabidopsis PHE1 AT1G65330 MADS-box transcription factor [96−99] Arabidopsis PLT2 AT1G51190 AP2/EREBP transcription factor [25] Arabidopsis RAN1 AT5G20010 Ras-related nuclear GTPase [34] RAN2 AT5G20020 RAN3 AT5G55190 Arabidopsis RAV1 AT1G13260 AP2/B3 domain transcription factor [76] Arabidopsis SHB1 AT4G25350 SYG1 family protein; transcription coactivator [20−22, 35] Arabidopsis TAR1 AT1G23320 Tryptophan aminotransferase; involved in auxin biosynthesis [38, 39] TAR2 AT4G24670 Arabidopsis TFL1 AT5G03840 Phosphatidylethanolamine binding protein [34] Arabidopsis WEI8 AT1G70560 Tryptophan aminotransferase; involved in auxin biosynthesis [38, 39] Maize ZmYUC1 Zm00001d023718 Flavin monooxygenase; involved in auxin biosynthesis [15, 47] Rice OsYUC11 Os12g08780 Flavin monooxygenase; involved in auxin biosynthesis [46] -
Transcription factors integrate developmental and environmental signals to play a central role in plant signal transduction. Many signaling regulators control development of the endosperm by modulating gene expression of HAIKU signaling components (Fig. 2). As these regulators are also involved in hormone signaling, the functions of these regulators are discussed in the 'Hormonal regulation of early endosperm development' section. This section mainly discusses the mechanisms of the HAIKU pathway during endosperm development.
The HAIKU pathway regulates endosperm proliferation and the timing of endosperm cellularization in Arabidopsis. This pathway comprises three core genes, HAIKU1 (IKU1), encoding a VQ-motif-containing protein, MINISEED3 (MINI3), encoding a WRKY transcription factor, and HAIKU2 (IKU2), encoding a leucine-rich repeat receptor-like kinase, which are expressed preferentially or specifically in early endosperm[17−19]. The haiku mutants iku1, mini3, and iku2 possess a small seed size phenotype without any distinguishable vegetable or other reproductive developmental defects[17,19]. The small seed size phenotype of the haiku mutants results from reduced growth of the endosperm and precocious endosperm cellularization, but the rate of free nuclei division is unaffected[17,19]. Cell elongation of the seed coat is reduced and embryonic cell proliferation is lower in the haiku mutants[17]. These characteristics of the haiku mutants make them ideal research objects for studying endosperm growth and communication between seed components. SHORT HYPOCOTYL UNDER BLUE1 (SHB1), encoding a transcriptional activator and containing an N-terminal SPX domain and a C-terminal EXS domain, promotes seed enlargement[20]. Seeds of the gain-of-function mutant shb1-D are larger than those of the wild type[20,21]. Genetic analyses have demonstrated that SHB1, MINI3, and IKU2 act in the same genetic pathway[20,21]. MINI3 binds directly to the W-BOXs of the MINI3 and IKU2 promoters and recruits SHB1 to activate their expression[20,21]. The abundance of the MINI3 transcript is also low in iku1 seeds, and the size of iku1 seeds is parallel to that of mini3 and iku2, indicating that IKU1 plays an important role in the HAIKU pathway[19]. It has been shown that IKU1 interacts with MINI3[18]. MINI3 may recruit IKU1 to activate the expression of MINI3 and IKU2 directly, or IKU1, MINI3, and SHB1 may form a transcriptional complex to regulate the expression of MINI3 and IKU2. Ectopic overexpression of AtSHB1 results in larger seeds in canola[22]. AtSHB1 directly regulates the expression of a set of key canola genes which are homologous to Arabidopsis MINI3, IKU2, SHB1, AGAMOUS-LIKE 62 (AGL62), and FERTILIZATION INDEPENDENT ENDOSPERM (FIE) in AtSHB1-overexpressing canola seeds. Consistently, SHB1 upregulates AGL62 and downregulates FIE directly in Arabidopsis[22]. These results indicate that SHB1 may modulate many aspects of seed development, including endosperm proliferation[17,20,21], endosperm cellularization[23], and FERTILIZATION-INDEPENDENT SEED-polycomb repressive complex 2 (FIS-PRC2) activity[24]. Two major regulators of plant cell totipotency, BABY BOOM (BBM) and PLETHORA2 (PLT2), act redundantly to promote proliferation of endosperm nuclei and endosperm cellularization in Arabidopsis[25]. BBM directly targets the FIE promoter[25]. BBM and PLT2 may redundantly repress FIE to regulate early endosperm development (Fig. 2). Wu et al. has demonstrated the evolutionarily conserved role of IKU2 during endosperm proliferation in dicot and monocot plants[26]. The expression of IKU2s is repressed epigenetically by the PRC2-mediated H3K27me3 in dicot plants, and is continuous during the syncytial and cellularized endosperm development due to the lack of H3K27me3 markers on IKU2s gene loci in monocot plants[26]. The authors proposed that the ancestral IKU2 function but divergent epigenetic regulation reveals the evolutionary route of seed development[26], shedding new light on the evolution of seed development from the perspective of endosperm development.
As mentioned above, HAIKU pathway regulates endosperm development through transcriptional regulation of IKU2. However, the molecular and biochemical functions of IKU2 remain largely unknown. Only one study has revealed that cytokinin biosynthesis and signaling act downstream of the HAIKU pathway[27]. Either IKU1, MINI3, or IKU2 activates the cytokinin degradation gene CYTOKININ OXIDASE 2 (CKX2)[27]. Cytokinin activity is higher in iku1 developing endosperm[27]. Genetic analyses have demonstrated that the small seed phenotype of iku2 is rescued in part by inhibiting cytokinin signaling or overexpressing CKX2 in endosperm[27]. MINI3 binds directly to the W-box of the CKX2 promoter and activates CKX2[27]. CKX2 expression is also suppressed by FIS-PRC2 in Arabidopsis[27]. These findings suggest that growth of the endosperm is coordinately regulated by genetics and epigenetics[27]. Notably, the expression of CKX2 is also downregulated in iku2 developing seeds[27]. However, IKU2 is not a transcription factor. Thus, activation of CKX2 by IKU2 is indirect (Fig. 1). Identifying the ligands and direct targets of IKU2 will elucidate the molecular and biochemical controlling mechanisms of IKU2-mediated seed size.
-
Post-transcriptional regulation participates in the control of endosperm-mediated seed size. Non-coding small RNAs have been demonstrated to regulate endosperm gene imprinting, which has been well reviewed[28]. Entry of sperm and/or pollen tube contents triggers central cell division and initiates development of the seed coat[29,30]. Sperm-transmitted miR159 promotes endosperm nuclear division by inhibiting central cell-transmitted MYB33 and MYB65, and ectopic expression of a miR159-resistant version of MYB33 in the endosperm suppresses the onset of endosperm nuclear division in Arabidopsis[31]. Zma-miR169o, which is highly expressed in the developing kernel, controls endosperm development and kernel size by regulating auxin biosynthesis[15]. The maternally expressed lncRNA MISSEN negatively regulates early endosperm development by competitively inhibiting the interaction between a helicase family protein (HeFP) and tubulin in rice[32]. In conclusion, the small RNAs and long non-coding RNAs are involved in the development of the endosperm and the control of seed size (Fig. 2).
-
Several studies have shown that early endosperm development involves post-translational regulation, including protein N-terminal acetylation, protein movement, and protein degradation (Fig. 2).
AGL62 is a type-I MADS transcription factor that is vital to proliferation and cellularization of the syncytial endosperm. The N-terminal acetyltransferase A (NatA) complex regulates protein N-terminal acetylation[33]. Mutations in Nα-acetyltransferase 10 (NAA10) and NAA15, the catalytic and auxiliary subunits of the Arabidopsis NatA complex, respectively, result in delayed and incomplete endosperm cellularization because of prolonged AGL62 expression[33]. More investigations are required to identify the NatA complex substrates during early seed development.
Three distinct mitotic domains are formed in the Arabidopsis syncytial endosperm[11]. A recent study revealed the precise control of protein movement between distinct syncytial endosperm regions. A mutation in TERMINAL FLOWER1 (TFL1) encoding a phosphatidylethanolamine binding protein causes larger seeds without changing the seed number per silique in Arabidopsis[34]. The TFL1 protein is produced in the chalazal endosperm and then moves to the cytoplasm of the syncytial peripheral endosperm by interacting with three small GTP-binding Ras-related nuclear (RAN) proteins, RAN1, RAN2, and RAN3. Genetic analyses have revealed that endosperm development regulated by TFL1 depends on functional RAN2. Because of the potential role of RANs in nucleocytoplasmic transport, it is necessary to investigate whether RANs regulate endosperm development through the nucleocytoplasmic transport process besides regulating TFL1 movement between two distinct endosperm regions.
Intriguingly, TFL1 interacts with ABSCISIC ACID INSENSITIVE 5 (ABI5) and affects its stability during seed development[34]. ABI5 suppresses endosperm proliferation by binding to the SHB1 promoter[35], suggesting that ABI5 localizes to the nucleus to regulate endosperm development. Previous studies indicated that ABI5 localizes in the nucleus and cytoplasm, which depends on the seed dormancy level and germination temperature in sunflower[36]. ABI5 is degraded via the ubiquitin-26S proteasome pathway in the cytoplasm under normal developmental conditions or in the absence of ABA[37]. TFL1 may interact with ABI5 in the cytoplasm of the syncytial endosperm to stabilize ABI5, indirectly leading to increased accumulation of ABI5 in the nuclei; thus, inhibiting endosperm proliferation.
-
Phytohormones are regulatory compounds that modulate plant growth and development. Previous studies suggested that auxin, cytokinin (CK), brassinolide (BR), ABA, and gibberellin (GA) play crucial roles in controlling seed size. However, the molecular mechanisms of the phytohormones during endosperm development are largely unknown.
Auxin
-
Auxin is produced rapidly in the Arabidopsis endosperm after fertilization[38]. Severe endosperm developmental defects have been observed in either auxin biosynthetic deficient mutant wei8/tar1/tar2 or auxin signaling deficient mutant axr1/axl[38]. These mutants display fewer, larger, and a disorganized distribution of endosperm nuclei, suggesting that auxin promotes early endosperm proliferation and is required for correct endosperm development (Fig. 2). The enlarged size of the nuclei in these mutants reflects endoreplication[38]. The excessive auxin in the Arabidopsis endosperm prevents syncytial endosperm cellularization, leading to aborted seeds[39,40]. It has been revealed that endosperm-derived auxin acts as a mobile molecular signal to trigger initiation of the seed coat[8,41]. Auxin is also involved in controlling proliferation of the integument cells and maternally regulates seed size in Arabidopsis[42].
Auxin biosynthesis and transport are regulated by AGL62 during early endosperm development. AGL62 expression is confined to the central cell and syncytial endosperm, and decreases abruptly just before cellularization in Arabidopsis[23,38]. Reduced endosperm nuclei proliferation, very precocious cellularization, and a failure to initiate the seed coat occur in agl62 mutant seeds[23,41,43]. The loss of FveAGL62 also results in the premature endosperm cellularization[44]. Auxin accumulation decreases severely in Atagl62 and Fveagl62 endosperm[44]. FveAGL62 interacts with FveAGL80 to form a heterodimer which suppresses several FveATHB genes, such as FveATHB29b and FveATHB30[44]. Many auxin biosynthetic genes are downregulated by FveATHB29b and FveATHB30 in the endosperm[44]. Overexpression of either FveATHB29b or FveATHB30 exhibits a similar phenotype to that of the Fveagl62 mutant[44]. AGL62 has also been proposed to regulate auxin transport from the endosperm to maternal sporophytic tissues in Arabidopsis and Fragaria vesca by modulating the expression of the potential auxin transporter-encoding genes in the endosperm[8,44]. Whether the AtAGL62 and FveAGL62 homologs regulate development of the syncytial endosperm by promoting auxin biosynthesis in monocot plants requires further confirmation (Fig. 2).
Many studies have focused on the functions of auxin during grain filling in monocot plants. The levels of auxin increase from 1 to 14 DAP, and the largest increase coincides with the start of the major starch storage phase in rice[45]. The OsYUC11 acts as a predominant contributor to auxin biosynthesis in endosperm during the grain filling stage. The expression of OsYUC11 is activated at 5 days after fertilization (DAF) and persistently increases before peaking at 15 DAF[46]. The Osyuc11 mutant displays a smaller seed and increased chalkiness[46]. The rice nuclear factor Y (NF-Y) protein OsNF-YB1 binds to the OsYUC11 promoter to induce OsYUC11 expression[46]. Mutations in OsNF-YB1 decrease indole-3-acetic acid (IAA) biosynthesis, leading to a smaller seed and increased chalkiness[46]. ZmYUC1 is highly expressed in the developing seed and plays a crucial role in the development of the maize endosperm[15]. The maize defective endosperm 18 (de18) mutant lacking the functional ZmYUC1 protein also exhibits impaired IAA biosynthesis in the endosperm, which results in reduced endosperm proliferation, endoreduplication, and kernel size[47]. According to these results, auxin promotes the proliferation of endosperm cells and enlarges the seeds in rice and maize (Fig. 2). Given that the transition from mitosis to endoreduplication is associated with a sharp increase in the auxin/CK ratio[15,48,49]. Early endosperm development may be regulated by the auxin-CK interactions. A recent study showed that auxin and CK coordinately regulate endosperm development in maize, reinforcing the hypothesis[15]. Overexpressing Zma-miR169o increases endosperm proliferation and seed size by negatively regulating its direct target ZmNF-YA13, which activates ZmYUC1 directly[15]. Overexpressing ZmNF-YA13 under control of the UBI promoter results in a slightly increased ZmYUC1 transcript level and auxin content but a significant increase in the auxin/CK ratio in developing kernels, likely leading to enlarged endosperm cells without rapid cell proliferation and reduced seed size[15]. The antagonistic action of auxin and CK has been demonstrated to regulate early embryo development[50]. Thus, early endosperm development is also under control of the interactions between auxin and CK, but this requires further confirmation.
Epigenetics is also involved in the regulation of auxin biosynthesis in the early endosperm. Many key auxin biosynthetic genes, such as OsYUC11, ZmYUC1, and AtYUC10, are paternally expressed in the endosperm by genomic imprinting[51]. The H3K27me3 markers are deposited on the AtYUC10 and AtTAR1 loci in the syncytial endosperm[52], indicating that FIS-PRC2, the major PRC2 in Arabidopsis endosperm, is involved in the direct regulation of auxin biosynthetic gene expression. FIS-PRC2 also directly inhibits non-imprinted AtAGL62, which plays a critical role in regulating auxin biosynthesis[23,40]. The expression of OsYUC12, which is transiently expressed during the syncytial endosperm development, and its close homologs OsYUC13 and OsYUC14 is inhibited by the rice polycomb group protein EMBRYONIC FLOWER2a (OsEMF2a) in the early endosperm[46,53]. Therefore, the development of the syncytial endosperm regulated by auxin and PRC2 might be evolutionarily conserved in dicot and monocot plants (Fig. 2). In addition, OsYUC11 is dynamically imprinted in rice endosperm[46], reflecting the complex nature of gene regulation during endosperm development.
In summary, auxin biosynthesis is coordinately regulated by genetics and epigenetics during early endosperm development; and auxin promotes endosperm proliferation and regulates normal nuclei distribution, the timing of cellularization, and grain filling.
CK
-
CK has been known to regulate endosperm development for decades and is used widely as a genetic target to increase crop yield potential; however, the molecular mechanism of CK-mediated endosperm development remains virtually unknown.
Similar to auxin, the CK level increases transiently in the endosperm after fertilization, which is correlated with the rapid division of the endosperm nuclei and cells[27,54]. In Arabidopsis, transcriptome analysis of syncytial stage endosperm indicates that CK signaling plays predominant roles in the development of the syncytial endosperm[55]. CK activity is low in unpollinated ovules and the seed coat[27]. The uniform and high CK activity is distinguished in the early syncytial endosperm. Then, CK activity decreases in the peripheral and micropylar endosperm and becomes restricted to the chalazal endosperm at the preglobular embryo stage in Arabidopsis[27]. Specific members of the gene families involved in CK biosynthesis and catabolism are expressed in opposite poles of the syncytial endosperm, likely leading to the formation of a gradient distribution of CK activity in the Arabidopsis syncytial endosperm[27,56]. The CK level in maize increases in the endosperm after fertilization and peaks at 10 DAP[57,58]. The gradient distribution of the CKs has also been detected in the developing kernel by CK immunolocalization[59]. The expression patterns of ZmIPT2 and ZmCKX1, which are the most likely gene family members controlling CK homeostasis in developing kernels, are also correlated with the spatiotemporal distribution of CKs[57−59].
It is challenge to access the precise functions of CK in the control of early endosperm-mediated seed size due to the pleiotropic effects of CK on determining the yield and the intricate nature of endosperm development[54]. It has been revealed that ZmIPT2 is subjected to artificial selection during the maize breeding, in which a single nucleotide change from cytosine (C) to thymine (T) in the ZmIPT2 coding region leads to an amino residue conversion[60]. The isopentenyl transferase activity of ZmIPT2-T is higher than that of ZmIPT2-C. The inbred lines carrying the ZmIPT2-T allele display higher kernel weight than those containing the ZmIPT2-C allele, suggesting that CK promotes growth of the endosperm and the enlargement of seeds. Most other studies have supported the hypothesis that endosperm growth, grain filling, and seed size are positively correlated with CK biosynthesis and negatively correlated with CK degradation[54]. However, the repression of CK activity by the HAIKU pathway is required for growth of the syncytial endosperm, suggesting that CK inhibits early endosperm growth and seed enlargement in Arabidopsis[27]. Arabidopsis histidine kinases (AHKs) act as cytokinin receptors and phosphorylate Arabidopsis histidine phosphotransfer proteins (AHPs) upon binding to CK[61]. The phosphorylated AHPs translocate to the nucleus where they phosphorylate Arabidopsis response regulators (ARRs) leading to the initiation or repression of gene transcription[61]. The ahk2/3/4, ahp2/3/5, arr1/10/12, as well as the CK signaling activator mutant cytokinin-independent 1 (cki1-8) all produce fewer yet larger seeds[61−64], reinforcing the hypothesis that CK suppresses seed enlargement. It has been proposed that sibling lethality has a strong effect on the size of the remaining seeds[65]. Thus, secondary and non-specific effects may contribute to the control of CK-mediated seed size in Arabidopsis. In the dicot tobacco, the modest increase in CK content by overexpressing the IPT gene under control of an endosperm-specific promoter, which is expressed from middle to late stages of seed development, produces larger seeds without any morphological abnormalities[66]. Given that tobacco seeds undergo ab initio cellular endosperm development and retain the endosperm to maturity[66], the development of the endosperm regulated by CK may be different in flowering species with distinct modes of endosperm development. It is also possible that the functional diversity of CKs during endosperm development derives from the developmental differences between the syncytial and cellularized phases (Fig. 2).
BR
-
BR and BR signaling regulate seed development in Arabidopsis and rice[67,68]. Seed size and shape mediated by BR largely depend on promoting cell division and expansion of the maternal tissues, which has been well reviewed[6]. BR activates the expression of SHB1, IKU1, MINI3, and IKU2 in Arabidopsis endosperm, which are positive regulators of endosperm proliferation[69] (Fig. 2). The BR-activated transcription factor BRASSINAZOLE-RESISTANT1 (BZR1) binds directly to the SHB1, IKU1, and IKU2 promoters[69]. The atypical non-DNA-binding basic-helix-loop-helix (bHLH) transcription factor ATBS1-INTERACTING FACTOR 2 (AIF2) acts downstream of BZR1 to negatively regulate seed enlargement[70]. Lower expression of SHB1, IKU1, and MINI3 transcripts is detected in developing seeds of AIF2 overexpressing plants[70] (Fig. 2). AIF2 expression is inhibited by BRASSINOSTEROID INSENSITIVE1 (BRI1)/BZR1-mediated BR signaling via direct binding of BZR1 to the AIF2 promoter in seedlings[71]. The non-DNA-binding bHLH transcription factor usually interacts with the DNA-binding bHLH transcription factor to antagonize its function. Future studies are required to identify which DNA-binding bHLH transcription factor interacts with AIF2 during seed development.
BR-deficient and insensitive mutants exhibit severe vegetative phenotypes, poor nutrient accumulation, and reproductive developmental defects. Overexpressing BR-biosynthetic gene in stems, leaves, and roots, but not embryos or the endosperms, also increases rice seed filling and yield, suggesting that BRs stimulate the flow of assimilate from the source to the sink[72]. The rice Epi-rav6 mutant, which is a gain-of-function epiallele of rice RELATED TO ABSCISIC ACID INSENSITIVE3 (ABI3)/VIVIPAROUS1 (VP1) (OsRAV6), activates BR signaling but develops smaller grains, which is different from the typical BR mutants[73]. These observations suggest secondary or indirect effects of BR on the control of seed size. Functional analysis of transgenic plants that specifically modulate BR signaling in the endosperm would be an efficient approach to uncover the regulation of endosperm development by BR.
ABA
-
In addition to its key roles in seed dormancy and germination, ABA regulates early endosperm development. The important ABA biosynthetic genes NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6 (NCED6) and ABA DEFICIENT 2 (ABA2) are highly expressed in the early Arabidopsis endosperm[35,74]. Expression of ABA-biosynthetic and signaling genes is not detected until 3 DAF in rice caryopses, and the ABA levels increase and peak at 6 DAF in the rice endosperm[75]. These results suggest that early endosperm development involves ABA signaling. ABA negatively regulates endosperm proliferation (Fig. 2). The seeds of the ABA-deficient mutant aba2 and the ABA-insensitive mutant abi5 are larger than those of wild-type Arabidopsis[35]. ABA inhibits endosperm proliferation through direct transcriptional repression of SHB1 by ABI5[35]. As mentioned above, accumulation of the ABI5 protein is mediated by TFL1[34]. RAV1 is a plant-specific B3 domain and AP2 domain-containing transcription factor that negatively regulates seed size by directly suppressing MINI3 and IKU2[76]. The expression of RAV1 is downregulated by ABA and BR in Arabidopsis seedlings[77]. BR-induced repression of RAV1 may be mediated by BZR1 which binds directly to the RAV1 promoter[76,78,79]. RAV1 directly suppresses ABI5 in Arabidopsis seedlings[80]. Further work is required to clarify whether these regulatory relationships between ABI5, BZR1, and RAV1 exist during early endosperm development (Fig. 2).
GA
-
The crucial roles of GA during seed dormancy and germination are well established. GA-deficient pea mutants exhibit smaller seeds compared with wild-type plants[81]. The levels of bioactive GAs are high during early endosperm development (6–8 DAP) but decrease significantly thereafter in maize endosperm[16]. A similar trend is observed in rice and barley seeds[82,83]. These results suggest a positive role for GA in the regulation of early endosperm growth (Fig. 2). DELLA, the key component of GA signaling, regulates cell expansion and division and suppresses plant growth and development[84]. GA triggers the degradation of DELLA through the ubiquin-26S proteasome pathway and promotes plant growth[84]. Two DELLA genes ZmGRAS54 and ZmGRAS12 are expressed at the early stage of endosperm development[16], suggesting that DELLA may regulate the expansion and/or division of early endosperm cells. In addition, the degradation of RGA in the integument is most likely triggered by auxin derived from the endosperm during the seed coat initiation process[8]. It is reasonable to say that GA signaling acts downstream of auxin during early endosperm development (Fig. 2).
-
Our understanding of the regulation of endosperm development by epigenetics is primarily based on investigations of imprinting which refers to the differential allelic expression of certain genes based on their parent of origin[28]. Genomic imprinting primarily occurs in the endosperm[28]. The mechanism of genomic imprinting involves DNA methylation, histone modifications, and activities of non-coding RNAs[28]. Histone acetylation and methylation are crucial for gene transcriptional regulation. Histone acetylation regulates development of the endosperm in maize[85−87]. Histone-modified H3K27me3 deposited by PRC2 is associated with transcriptional repression and plays an important role during early endosperm development (Fig. 2).
Three major PRC2s have been described in Arabidopsis: FIS-PRC2, EMF-PRC2, and VRN-PRC2[24]. During early seed development, FIS-PRC2 mainly regulates endosperm and embryo development; EMF-PRC2 and VRN-PRC2 mainly control the development of sporophytic tissues[24,88]. Four subunits, FERTILIZATION INDEPENDENT SEED 2 (FIS2), FIS1/MEDEA (MEA), MULTI-COPY SUPPRESSOR OF IRA1 (MSI1), and FIE, compose the core FIS-PRC2[24]. Mutations in these genes decrease the accumulation of H3K27me3, resulting in autonomous seed development in the absence of fertilization and aborted seeds due to excessive endosperm nuclear proliferation, failure of cellularization, and embryo arrest after fertilization[24,88]. FIS-PRC2 appears to be Brassicaceae-specific, as the AtFIS2 and AtMEA homologs are only found in Brassicaceae[24]. However, PRC2 is also involved in early endosperm development in other plant species[24,89]. Rice has two AtFIE homologs OsFIE1 and OsFIE2 which are all involved in grain weight control[90−93]. Intriguingly, OsFIE1 has been reported to regulate seed size under heat stress[94]. Of the AtSWN-like genes in soybean, Gm11G067000 rescues the Atmea-3 seed lethality phenotype and is considered as the GmMEA gene[26]. OsEMF2a plays a similar role as AtFIS2 during genomic imprinting and early endosperm development in rice[53,95]. The Osemf2a mutants display autonomous endosperm proliferation in the absence of fertilization and delayed cellularization after fertilization[53,95]. OsEMF2a also regulates the expression of the type Ⅰ MADS-box genes which might be involved in early endosperm development[53,95]. These findings reflect the evolutionarily conserved roles of PRC2 during early endosperm development in dicot and monocot plants.
Many imprinted and non-imprinted genes are regulated by FIS-PRC2 in Arabidopsis[52]. As mentioned above, FIS-PRC2 regulates the expression of hormone biosynthetic genes and IKU2 during early endosperm development (Fig. 2). FIS-PRC2 also represses transcriptional abundance of the core transcription factors which are crucial for early endosperm development (Fig. 2). The AGL62 expression pattern may be vital for maintaining the normal auxin dynamics which contribute to precisely regulate endosperm proliferation, cellularization, and seed coat initiation. Although AGL62 is not an imprinted gene, FIS-PRC2 directly inhibits AGL62 in the endosperm[23,40]. FIS-PRC2 may be recruited by particular transcription factors expressed immediately before cellularization to the AGL62 locus. FIS-PRC2 silences the maternal PHERES1 (PHE1) allele after fertilization[96]. PHE1 is proposed to act as a transcriptional activator and may directly activate YUC10, MINI3, IKU2, and AGL62 in the endosperm[97,98]. The phe1 mutant has no abnormal seed phenotype, probably due to functional redundancy with other genes[97,98]. The type I MADS-box transcription factors usually form heterodimers to increase DNA binding specificity[23,44,99]. PHE1 interacts with AGL28, AGL40, and AGL62 in yeast[99]. Thus, identifying the PHE1 redundant genes and the PHE1 interacting proteins during endosperm development would enrich our understanding of the regulatory mechanism of endosperm development. FIS-PRC2 also silences expression of the important seed maturation gene FUSCA3 (FUS3) during early endosperm development[100]. The BASIC PENTACYSTEINE (BPC) proteins BPC1 and BPC2 directly bind to the FUS3 promoter and suppress its expression by directly recruiting FIS-PRC2[100]. The ectopic expression of FUS3 in bpc1/2 endosperm results in over-proliferation of the endosperm, causing delayed or arrested embryonic development[100]. Thus, precise spatiotemporal gene expression regulated by PRC2 during early endosperm development is crucial for normal endosperm and embryo development (Fig. 2).
-
Although the early endosperm developmental period is short, it is critical to seed development. The proliferation of syncytial endosperm nuclei and the endosperm cell division that occur during early endosperm development play a crucial role in determining seed size. Substantial progress has been made in the molecular regulation of endosperm development in recent decades. However, the molecular mechanisms of early endosperm development are not clearly understood. Many important questions remain unsolved. The biochemical function of the receptor kinase IKU2 is a major puzzle in HAIKU signaling. The exact molecular functions of hormones during endosperm development and the control of seed size require additional investigations, due to the problematic pleiotropic nature of hormones. Hormonal interactions may be involved in endosperm development, such as auxin-CK interactions, ABA-BR interactions, and auxin-GA interactions. Epigenetics is involved in genome-wide regulation of imprinted and non-imprinted gene expression during early endosperm development. However, our understanding of the mechanisms of specific gene expression regulated by epigenetics during endosperm development is very limited. Biochemical approaches and new technologies, such as the single-cell/nuclear sequencing and genome editing, will be crucial for gaining clearer insight into the endosperm biology. Ultimately, a comprehensive understanding of the regulatory mechanisms of early endosperm development from basic studies will provide new guidelines and strategies to improve crops.
This research was funded by the key program of the National Natural Science Foundation of China (31730008).
-
Xiansheng Zhang is the Editorial Board member of journal Seed Biology. He is blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research group.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xu G, Zhang X. 2023. Mechanisms controlling seed size by early endosperm development. Seed Biology 2:1 doi: 10.48130/SeedBio-2023-0001
Mechanisms controlling seed size by early endosperm development
- Received: 26 October 2022
- Accepted: 19 December 2022
- Published online: 30 January 2023
Abstract: Seed size is an important agronomic trait determining the yield potential of crops that is controlled by the growth and development of the endosperm, embryo, and seed coat. The seed coat and endosperm have been proposed to play primary roles in determining seed size. Extensive research has been carried out on the regulation of seed size by seed coat, whereas the molecular mechanism underlying the regulation of seed size by the endosperm is poorly understood. Recent studies have emphasized the central role of the endosperm in seed development. The proliferation of syncytial endosperm nuclei and the endosperm cell division during early endosperm development determine the number of endosperm cells, which plays a fundamental role in controlling seed size. Here, we summarize the recent progress in early endosperm development, emphasizing the roles and molecular mechanisms of the HAIKU pathway, phytohormones, and polycomb repressive complex 2 in the control of seed size.
-
Key words:
- Endosperm development /
- Seed size /
- HAIKU pathway /
- Phytohormones /
- PRC2













