-
In plant science, seed is a product of sexual reproduction. Upon pollination, a pollen tube delivers two sperm cells to the embryo sac, one sperm cell fuses with the egg cell and the other fuses with the central cell, thus, triggering seed development. The fertilized egg cell, the zygote, develops into an embryo, while the fertilized central cell, the primary endosperm cell, divides to form the endosperm. Although plenty of diversities on morphology and structure exist in different species of angiosperms, generally a seed includes three major structures, embryo, endosperm and seed coat derived from the integuments of the ovule (Fig. 1). In some species, the established endosperm undergoes a programmed cell death (PCD) process and degenerates during seed maturation.
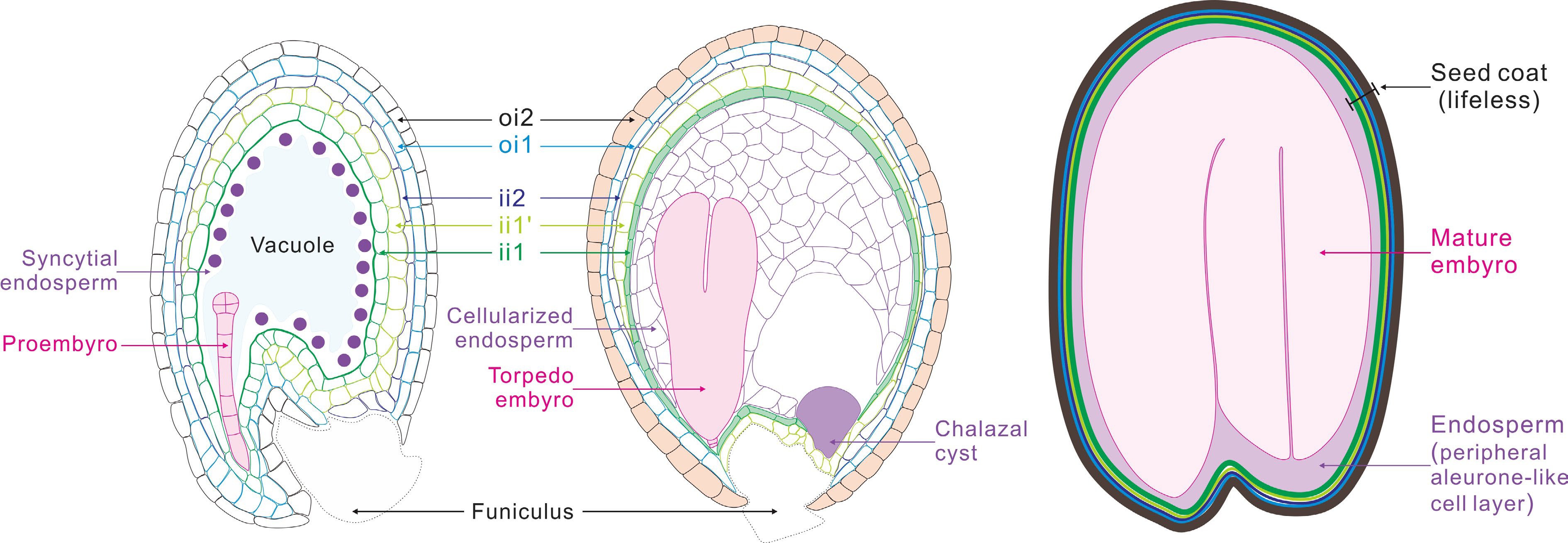
Figure 1.
Schematic representation of seed development in Arabidopsis. After fertilization, the fertilized egg cell (zygote), develops into a proembryo, which then undergoes cell differentiation and organ formation to produce a mature embryo. The primary endosperm cell undertakes continuous mitosis without cell wall formation to form the syncytial endosperm. Then, the endosperm coenocyte initiates cellularization to produce the cellularized endosperm. Finally, the endosperm undergoes programmed cell death (PCD), resulting in only one layer of endosperm cells wrapping the embryo in mature seeds. The two cell layers of outer integument (oi) and three cell layers of inner integument (ii) undergo a period of growth after fertilization, and then adopt distinct cell fates. The innermost endothelial layer (ii1) synthesizes the flavonoid compound proanthocyanidins, and the outermost epidermal layer (oi2) produces and secretes mucilage. By seed maturity, cells of all ii and oi layers are dead.
The seed has always been a key factor in agricultural production. Modern crop production and the science of agriculture confirm that, without seeds, successful agricultural production will not be possible as seeds are the cornerstone of agriculture as they are the propagules that insure plant genetic survival from one generation to the next. As the most important food, seeds have also been a central topic in plant science for decades. Accumulated information greatly enhanced our understanding of the different seed types, seed structures, and the processes of seed development, especially the molecular mechanisms regulating these processes in more recent decades. These new insights into seed development provide a powerful knowledge source and useful tools for crop improvement in both seed quality and yield. New technologies, e.g. genome editing, single cell RNA-seq, notably accelerated the research of seed biology and promote seed engineering for apomictic seeds, medicinal seeds and special nutritional seeds.
Arabidopsis has been used for seed developmental research since it was suggested as a useful model plant and has contributed to the advances in this field. Because of its small genome, short growth cycle, convenient planting and other advantages, Arabidopsis is rapidly and widely used in plant development research, which provides plenty of the genetic, cytological and development biological information. More importantly for seed biology research, Arabidopsis has the following advantages: 1) a large number of relatively synchronized developed seeds produced per silique; 2) large number of seeds produced for each pollination, thus easy for genetic studies; 3) easy to clear for the examination of embryo and endosperm development; 4) traceable patterns for embryo and endosperm development. Thus, it has been a major model plant for the investigations of seed biology. These investigations not only enhanced our understanding of the mechanisms regulating seed development in dicots but also accelerated relevant research in monocots, especially in crops.
In this brief review we try to summarize what we have learned in past 30 years on seed development. However, we realized that it is impossible to include all the findings in this review due to space limitation. We have to focus on some aspects of seed development and emphasize critical questions dealing with embryogenesis, endosperm development, and seed coat formation. Hopefully, the present review can provide an outline of the major advances in the research field of seed development.
-
In flowering plants, fertilization marks the initiation of a new sporophyte generation, which starts from the fusion of a haploid female gamete, the egg cell, and a male gamete, the sperm cell, leading to the formation of a diploid zygote[1]. Our current understanding of plant embryogenesis is mainly derived from the research regarding the model eudicot plant Arabidopsis. The whole process of embryo development is usually divided into two major periods: embryonic morphogenesis and embryo maturation[2,3]. In the first period, embryo development is characterized by a series of sequential embryo developmental stages based on the morphology of the embryo, during which the basic embryo body is established. Whereas, in the maturation process, embryos accumulate different storage materials and enter into a dormant state with the loss of water content.
In Arabidopsis, the zygotic genome is soon activated to synthesize novel transcripts after fertilization, which is accompanied by the clearance of a significant fraction of maternal transcripts stored in the egg cell. Then, the fertilized egg cell elongates rapidly and divides asymmetrically to form a small apical cell and a larger vacuolated basal cell. The two daughter cells of the zygote display different morphologic characteristics and distinct developmental fates during embryogenesis[4−6] (Fig. 2). The smaller apical cell divides to form the major parts of a mature embryo through a series of elaborate developmental events including the establishment of apical-basal axis and radial embryo pattern, while the larger vacuolated basal cell undergoes several rounds of cell divisions to form a 7–9-celled suspensor. The uppermost suspensor cell will develop into the hypophysis, and other suspensor cells will degenerate at later stages through PCD[7,8]. In this review, we will give an overview of some critical developmental events, including zygotic genome activation (ZGA), zygote development, cell fate determination, embryo pattern formation, based on the studies of Arabidopsis embryogenesis and embryo maturation over the past 30 years.
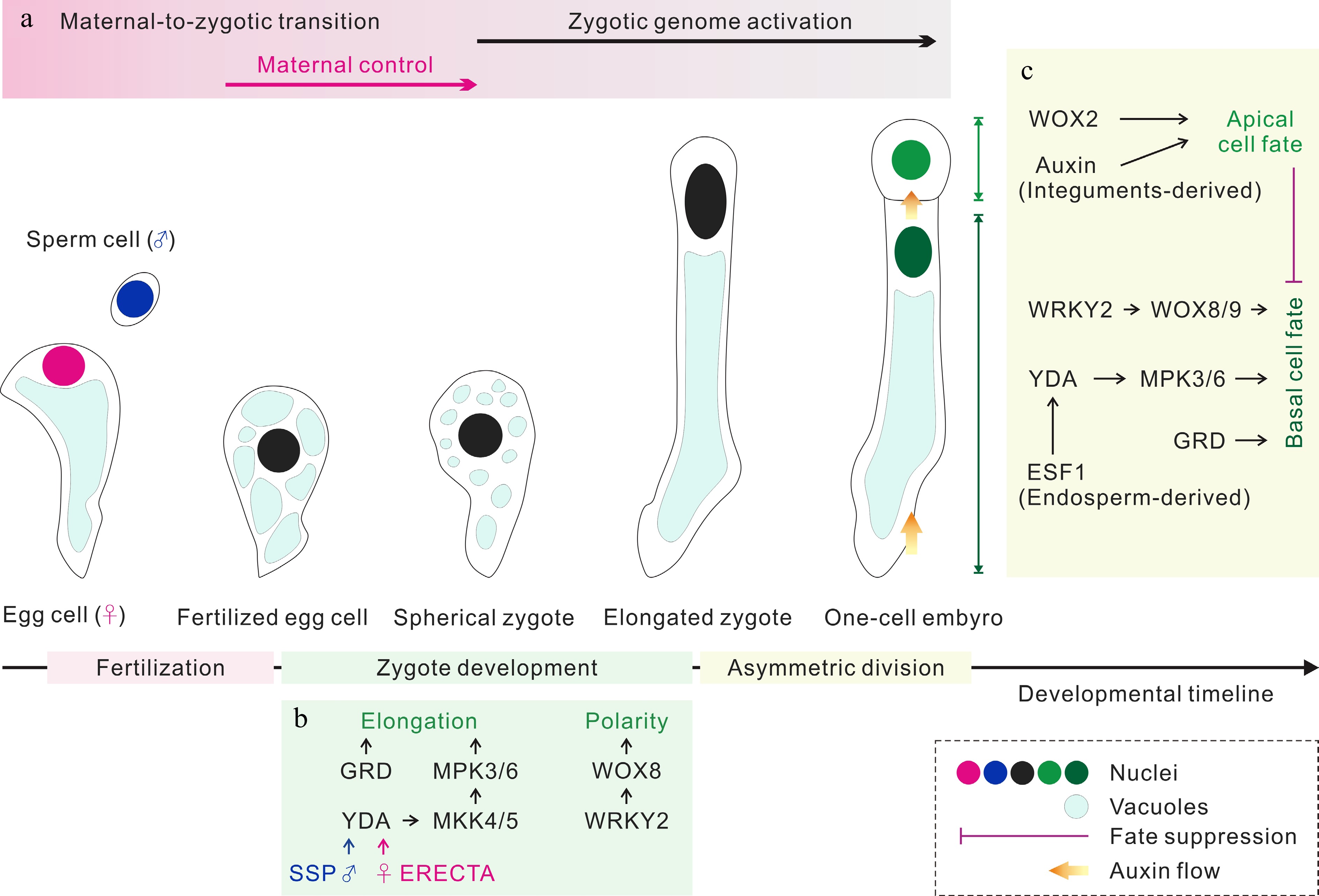
Figure 2.
Zygote development and apical/basal cell fate determination. (a) The parental contributions to the transcriptomes of early embryos. (b) The regulation of zygote development. Both sperm cell-derived SSP and egg cell-provided ERECTA trigger the YDA MAP kinase signaling pathway, which together with RWP-RK Factor GRD to elongate zygote. WRKY2 activates WOX8 to induce the polarity establishment of zygote. (c) The regulation of apical/basal cell fate determination. Following asymmetric zygote division, the auxin response and WOX2 control the apical cell fate, and the WRKY2-WOX8/9 and YDA MAP kinase pathways regulate the basal cell fate. GRD and the endosperm-derived ESF1 regulate basal cell lineage specification through the YDA MAP kinase pathway. In addition, the embryogenic potential of the basal cell lineage is suppressed by the apical cell lineage during normal embryogenesis.
Early zygotic genome activation (ZGA)
-
In most animals, early embryo development is controlled by maternal factors deposited in the egg cell, ZGA starts after one to several cell cycles in different species[9,10]. Although the research on ZGA was also extended to Arabidopsis about 20 years ago[11,12], the picture regarding the timing and scale of ZGA in Arabidopsis did not become clear until recently. The transcription activity in early embryos was investigated using LhG4/pOp transactivation system. The results revealed that the Arabidopsis zygotic genome is not silenced, and both maternal and paternal genomes are transcriptionally active[13]. The transcriptional activities in the egg cell and zygotes were also visualized by immunostaining of phosphorylated serine 2 on the carboxy-terminal domain of RNA polymerase II (RNAPII Ser2P). The RNAPII Ser2P signal in zygotes was higher than that in the egg cell, indicating that increased transcriptional activities occur shortly after fertilization[14].
Recently, comparative analysis of the transcriptome of Arabidopsis egg cells and zygotes revealed that the transcriptome was reconstructed shortly after fertilization and the zygote showed a distinct transcriptome compared with the egg cell on a genome-wide scale[5]. Accordingly, 2,625 genes and 2,951 genes were upregulated in the zygote at 14 and 24 h after pollination (HAP), implying 13.7% of genes in the Arabidopsis genome were transcribed to generate novel transcripts. These upregulated genes in early zygote were enriched in DNA transcription and RNA biosynthesis related pathways, while upregulated genes in elongated zygote were enriched in cell division related pathways, such as cell cycle and chromosome organization[5], suggesting that ZGA occurs with two successive activation waves at the zygotic stage. In addition, early Arabidopsis zygotes treated with the transcriptional inhibitor α-amanitin failed to elongate and divide[5,14]. All this evidences implies that, in contrast to animals, ZGA occurs shortly after fertilization to generate novel transcripts, which are required for zygote elongation and asymmetric division in Arabidopsis (Fig. 2a).
Zygotic polarity establishment
-
In Arabidopsis, egg cells usually displays obvious polarity with a nucleus at the chalazal end and a large vacuole at the micropylar end. After fertilization, fertilized egg cells will then undergo a series of morphological changes including the position of the nucleus, the organization of the cytoskeleton, the morphology and distribution of vacuoles to form an elongated zygote[4−6]. During this process, fertilized egg cell undergoes two critical developmental stages. In the first stage, the original large vacuole derived from the egg cell disappears rapidly and forms numerous small vacuoles, resulting in the formation of a spherical zygote without obvious polarity. The spherical zygote will then elongate rapidly in the second stage to form an elongated zygote with about 2.8- fold cell length compared with the fertilized egg cell[5,6]. During this elongation process, the zygote will rebuild its polarity, accompanied by the movement of the nucleus to the apical pole and the formation of a large vacuole at the basal pole[5,6] (Fig. 2). Besides the changes of nucleus position and vacuole morphology, rearrangement of the cytoskeleton is also occurred during the process of polarity establishment. Before fertilization, microtubules display a longitudinal array and F-actin shows a mesh-like structure in the egg cell. After fertilization, cytoskeleton loses its original structure, and reorganizes into a new pattern for supporting zygote development. Microtubules will develop into a transverse ring at the subapical region of a zygote, which is critical for zygote elongation, whereas F-actin forms an apical cap and longitudinal arrays, which is required for the movement of the nucleus to the apical end[4].
Molecular components in MITOGEN-ACTIVATING PROTEIN (MAP) kinase pathway including MAPKK kinase YODA (YDA), the MAPKs MPK3 and MPK6, and the MAPKKs MKK4 and MKK5 have been well characterized in regulating zygote polarity establishment[15,16]. In the yda mutant, the zygote could not properly elongate and led to an almost symmetric division[16]. YDA activity is activated by a membrane associated pseudokinase SHORT SUSPENSOR (SSP), which controls zygote development through a parent-of-origin effect[17]. In addition to SSP, its two paralogs BSK1 and BSK2 also lie upstream of the YDA signaling pathway, and act in parallel to SSP to regulate zygote elongation[18]. Recently, the receptor kinase ERECTA was also reported to be involved in zygote polarization as a maternal component of the YDA signaling pathway[19], suggesting the integration of paternally regulated SSP-YDA signaling pathway and maternally contributed ERECTA-YDA in zygote polarization (Fig. 2b).
In addition to the YDA signaling pathway, zinc-finger transcription factor WRKY2 was also shown to be required for zygote repolarization from the symmetric state[20]. The nucleus of 23.1% wrky2 zygote could not locate at the apical end unlike that of the wild type (WT) plants, and the vacuoles distribute throughout the zygote. Either maternally or paternally derived WRKY2 could rescue the defects of wrky2 zygote. Zygotes from the reciprocal crosses between wrky2 and WT plants are indistinguishable from the WT zygotes. The function of WRKY2 in zygote repolarization is dependent on its downstream target WOX8. Expression of WOX8-YFP in the wrky2 mutant significantly complements the defects of wrky2 in zygotic polarity establishment. However, wrky2 egg cells show similar morphological characteristics to the WT egg cell with a nucleus at the apical region and a larger vacuole at the basal end, suggesting that polarity establishment of egg cell and zygote are not coupled with each other, and likely regulated independently[20] (Fig. 2b). The interaction between MAP kinase signaling pathway and WRKY2-WOX8 pathway was dissected in a recent report. MAP kinase signaling cascade phosphorylates WRKY2 and activates its expression[21], revealing how paternal YDA signal is integrated in the regulation of zygote polarization.
Asymmetric zygote division
-
Zygotic division is the first crucial cell division during early plant embryogenesis, In Arabidopsis, the zygote usually divides asymmetrically to form a smaller apical cell and a larger basal cell. These two daughter cells display distinct cell division patterns and developmental fates. The smaller apical cell will develop into the main body of the embryo, while the larger basal cell will develop into the suspensor through limited cell divisions[2]. In the past 30 years, great efforts have been made to elucidate two fundamental aspects during zygote division: the initiation of zygote division and the regulation of asymmetric zygote division. Genome wide screening of zygote-arrested mutants has prompted research on the molecular mechanisms regulating the initiation of zygote division. A series of zygote-arrested mutants including fac1, fac19, zeus1, zyg1, zyg3 and gcd1 have been identified[22−26]. In addition to the similar zygote-lethal phenotype, all these mutants except gcd1 are recessive mutations. Furthermore, the function of parental alleles of FAC1, FAC19 and ZYG3 in zygote division has been tested[22,25−27]. During the reciprocal cross between the WT plants and these three zygote-arrested mutants, either the WT maternal allele or the WT paternal allele could complement the zygote-lethal phenotype, suggesting that early action of paternal genomes in early embryogenesis, and both maternal and paternal genomes contribute to the initiation of first zygote division. The research on GCD1 reveals another different mechanism underlying the initiation of zygote division. Mutant gcd1 egg cells could be fertilized with the WT sperm cell, but could not initiate fertilized egg cell division, suggesting egg cell deposited information is also crucial for zygote division[24]. All these data suggest that both gametic and zygotic transcripts are critical for the initiation of zygote division, although pivotal paternal factors responsible for triggering fertilized egg cell division are still unknown in Arabidopsis.
For understanding the regulation of asymmetric zygote division, Arabidopsis mutants with near symmetrical zygote division have prompted investigations into the establishment of zygotic division plane and the effect of different zygote division manner on cell lineage specification. As discussed above, the YDA signaling cascade is critical not only for the zygote polarization, but also for the asymmetric division. For example, ssp and yda zygotes failed to elongate and both divided symmetrically, and the basal cells could not differentiate into a recognizable suspensor[16,17]. GROUNDED (GRD), encoding a putative RWP-RK family transcription factor, is required for YDA-dependent signaling in the zygote. grd mutant is defective in zygotic cell elongation and shows abnormal zygote division patterns[28,29]. These data suggest that the asymmetric zygote division seems critical for subsequent embryonic pattern formation, especially for the basal cell lineage differentiation. However, the defects in zygote elongation are coupled with the symmetrical zygote division in all these mutants. Rapid elongation of the zygote seems independent to the initiation of zygote division, but related to zygotic division patterns. Whether the defects in embryonic pattern formation are due to the defects in zygote elongation or the symmetrical zygote division remains to be further clarified.
In addition to the YDA signaling pathway, several other factors such as GNOM, FASS and ZAR1 are also involved in the asymmetric zygote division[30−32]. GNOM encodes a guanine nucleotide exchange factor for ADP-ribosylation factor, and thus has potential roles in vesicle trafficking. Loss of GNOM results in the formation of a lesser-elongated zygote, which then divides symmetrically to form two daughter cells with nearly equal size[30]. The apical daughter cell then undergoes abnormal cell division, which results in an octant embryo without typical apical-basal pattern, suggesting that the zygote asymmetric division is directly or indirectly linked to subsequent apical-basal pattern establishment. Since GNOM is required for the polar localization of PIN1[33], the role of auxin in regulating asymmetric zygote division is worthy of investigation. Whereas in fass mutants, zygotic polarity seemed unaffected, but the orientation of zygotic cell division was impaired[32]. Recently, research on RLK/Pelle kinase ZAR1 provides us an opportunity to understand the effect of zygote polarity and asymmetrical division on subsequent apical and basal cell fate specification. ZAR1 mutation does not impair zygote elongation but disrupts its asymmetric division pattern. And the developmental fate of both apical and basal cells is impaired, suggesting that zygote intrinsic asymmetrical division pattern is likely critical for cell fate determination during embryogenesis[31]. In addition, ZAR1 could interact with calmodulin CaM1 and the heterotrimeric G protein Gβ, indicating that ZAR1 acts as an integrator for Ca2+ and G protein signaling to regulate zygote division. It is worthy to further study Ca2+ and G protein signaling cascades in asymmetric zygote division in the future.
Cell fate determination of apical and basal cell lineages
-
Cell fate determination is critical for embryonic pattern formation and morphogenesis during embryogenesis. Asymmetric zygote division generates two daughter cells with distinct development fates. Three potential molecular mechanisms have been used to explain the distinct development cell fates of apical cells and basal cells: 1) apical and basal cell fate may be determined by its relative position in the embryo sac, which are conferred by extrinsic environmental cues from maternal tissues; 2) cell fates of two daughter cells may be determined by asymmetrical division of the zygote, which results in uneven distribution of cytoplasmic factors related to cell fate determination; 3) cell fate determination may be controlled by the communication between apical and basal cell lineages[34−36]. During past decades, great advances have been made in the understanding of cell fate determination of apical and basal cell lineages during early embryogenesis.
Since suspensor connects the embryo to the surrounding endosperm and seed coats, it has long been proposed that extrinsic signals from endosperm and seed coats are likely involved in cell fate determination of basal cell linage. There are two examples supporting this proposal. Firstly, endosperm-expressed EMBRYO SURROUNDING FACTOR 1 (ESF1) peptides were found to be essential for the basal cell lineage specification. ESF1 peptides are exclusively expressed in the endosperm, and promote basal cell lineage specification through the YDA signalling pathway in a non-cell-autonomous manner[37]. Another example is that integument generated auxin signals are important for the correct embryo pattern formation (Fig. 2c). Although cell fate of apical and basal cell lineages has not been investigated in detail, abnormal cell division pattern were observed in both apical and basal cell lineages of wei8 proembryos[38], suggesting that maternal derived auxin seems critical for cell fate specification of early embryos.
For the second proposal, comparison of gene expression profile between apical and basal cells is an efficient approach to investigate the intrinsic factors for apical and basal cell lineage specification. Comprehensive studies about WOX (WUSCHEL related homeobox) family genes have helped us to understand the mechanisms underlying apical and basal cell fate specification. In Arabidopsis, two WOX family genes, WOX2 and WOX8, were reported to be involved in apical cell and basal cell fate determination. WOX2 and WOX8 were co-expressed in the zygote, and became confined in apical and basal cells after the asymmetric division, respectively[35]. In the subsequent embryo developmental process, WOX2 was mainly involved in embryonic shoot patterning. In wox2 mutant, nearly one third of embryos displayed abnormal periclinal divisions at the 8-cell embryo stage. In addition to WOX2, concomitant loss of other apical WOX genes resulted in the formation of shoot-less structures[39]. Although no visible defect was observed in wox8 mutant, embryo pattern formation in wox8 wox9 double mutants was severely disrupted[39] (Fig. 2c). Recently, by overcoming technique limitations, apical and basal cells, as well as embryo propers and suspensors of 32-cell embryos were successfully isolated and collected. Then the cell lineage specific transcriptomes were constructed. Comparative transcriptome analysis revealed that distinct transcriptomes were established in apical and basal cells immediately after zygotic division. Significant upregulation of 3,454 genes was found in the apical cell when compared with the basal cell, and 2,911 genes were significantly downregulated in the apical cell. The differences between the transcriptomes of apical and basal cell lineages were enlarged as embryos developed. Embryo-related pathways, such as DNA replication and embryo development, were specifically activated in apical cell lineage, whereas, suspensor-specific pathways including transportation and PCD are activated in basal cell lineage as early as 1-cell embryo stage. Although exact roles of long noncoding RNAs (lncRNAs) in plant cell fate determination are still largely unknown, in addition to protein coding genes, hundreds of differentially expressed lncRNAs between apical and basal lineages have also been identified. Elucidating the roles of these lineage-specific lncRNAs will enhance our understanding on the mechanisms of cell fate determination during early embryogenesis[40]. All these data suggest that cell lineage specific transcripts likely contribute to cell fate determination during early embryogenesis.
Besides cell type specific genes, MAP kinase cascade including YDA and SSP is also involved in regulating the specification of basal cell lineage. In yda mutants, zygotes failed to elongate and divided to form a WT-like apical cell and a smaller basal cell compared with that of WT proembryos. The apical cell displayed a similar division pattern as that in WT plants before 8-cell embryo stage, whereas the basal cell exhibited an abnormal division pattern and failed to differentiate into the suspensor. Correspondingly, gain-of function of YDA led to exaggerated growth of the basal cell lineage[16]. Similar to the yda, no typical suspensor was formed in the ssp mutant, and suspensor-specific marker SUC3 was not detected in the ssp proembryos, supporting the role of SSP in regulating the cell fate specification of basal cell lineage[17]. Since zygote elongation and division patterns were impaired in both yda and ssp mutants, whether basal cell fate specification is likely linked to the zygote elongation and asymmetric division. In addition to MAPK cascade, RWP-RK Factor GROUNDED (GRD) was also shown to regulate cell fate specification of basal cell lineage through YDA MAP Kinase Signaling (Fig. 2c). Mutations of GRD resulted in partial loss of asymmetric zygote division and disappearance of the expression of suspensor marker gene WOX8 in basal cell lineage[29,41]. Since GRD is co-expressed with YDA in the apical and basal cell lineages, how its activity is only restricted to basal cell fate specification is still poorly understood.
There are also numerous evidence supporting the third proposal that the communication between apical and basal cell lineages also contributes to the cell fate determination. This proposal was originally derived from the phenotype of several Arabidopsis mutants such as sus, twn, raspberry, iyo and rpl18aB[42−46]. In these mutants, a common characteristic is that suspensor cells did not initiate PCD, but continue to divide to form another embryo-like structure, suggesting that the basal cell lineage also has the embryogenic capacities to differentiate into an embryo as the apical cell lineage. In addition, the embryogenic potential of suspensor cells could be induced by expressing several known embryogenesis-inducing genes including RKD1, RKD4 and WUS in the suspensor cell, but the expression of other known embryogenesis-inducing genes such as BBM, LEC1 and SERK1 could not induce the cell fate transition from the suspensor into the embryo, indicating that the embryogenic potential of the suspensor could be triggered by confined reprogramming pathways[47]. In addition to expressing embryogenesis-inducing genes in the basal cell lineage, removing the interaction between apical and basal cell lineages or destroying the apical cell at the 1-cell embryo stage by in vivo living cell laser ablation technique could also promote the transition of the suspensor cells into an embryo, providing direct evidence for the fact that the embryogenic potential of the basal cell lineage is suppressed by the apical cell linage during normal embryogenesis[48,49] (Fig. 2c). However, the embryogenic potential of basal cell lineage is dependent on the embryo developmental stages. Suspensor will lose its embryogenic potential after the globular embryo stage, probably after the initiation of PCD[48]. To date, which signal from the apical cell linage suppresses the embryogenic potential of basal cell lineage is still unknown. Auxin is believed to be involved in the cell fate transition of basal cell lineage. Different auxin response components were found in apical and basal cell lineages, enabling different auxin responses for embryo and suspensor specification. In the basal cell lineage, ARF9/13 and IAA10 are the main components of auxin response machinery. Inhibition of auxin response in the basal cell lineage resulted in the loss of suspensor fate, and partial transition into embryo fate[50]. Recently, an auxin-dependent basic Helix Loop Helix transcriptional module was identified as the embryo development in the suspensor by mediating the auxin activity[51]. Further confirming the role of auxin-dependent transcription program is critical for understanding the cell fate specification of basal cell lineage.
On the other hand, there is multiple evidence supporting that basal cell lineage is also critical for apical cell lineage development. The first evidence that supports this notion is the studies from auxin efflux regulator PIN-FORMED 7 (PIN 7), which is located apically in the basal cell and mediates the apical-basal directional auxin transport. This auxin activity gradient is required for apical cell lineage development, suggesting that basal cell lineage may be required for embryonic pattern formation[52]. dAberrant cell division was observed in the apical cell lineage when removing the basal cell using laser irradiation[49]. All these data suggest that the interaction between basal cell linage and apical cell linage is critical for both suspensor formation and embryo development during early embryogenesis.
Parental contributions to early embryogenesis
-
Over the past 30 years, different approaches including reporter line analysis, genetics analysis of EMBRYODEFECTIVE (EMB) genes and transcriptome analysis of hybrid embryos have been used to investigate parental contributions to early plant embryogenesis. Two early studies led to controversial conclusions about paternal genome activation in Arabidopsis embryos. In the first study, authors demonstrated that paternal alleles of 20 loci in the Arabidopsis genome were silenced in early embryos and activated three to four days after fertilization[11]. In the second study, paternal alleles of three other genes including RPS5A, KNOLLE and KEULE were shown to be active in early embryos[12]. Transcriptome studies have also been used to investigate the paternal genome activation and parental contributions to the transcriptome of early embryos. Autran et al. demonstrated that the transcriptome of hybrid 2-4 cell and globular embryos derived from the cross between Landsberg erecta (Ler) and Columbia (Col-0) was dominated by maternal transcripts[53]. Whereas Nodine et al. showed equal parental contributions to the transcriptome of early embryos derived from the reciprocal crosses between Col-0 and Cvi-0[13]. The different conclusions from these early reports were explained by two potential theories, the contamination of transcriptomes by endosperms and seed coats or the different hybrid Arabidopsis ecotype combinations in two studies[54,55]. Recent transcriptomic analysis of hybrid Arabidopsis zygotes and early embryos from the reciprocal cross between Col-0 and Ler demonstrated again equal parental contributions to the transcriptomes of early embryos[5,56]. Interestingly, parental contributions to the transcriptome of zygotes display a developmental stage dependent manner. The transcriptome of very early zygote is dominated by maternal transcripts, which are due to the delivery of maternal transcripts from the egg cell into the zygote upon fertilization, but not due to the preferential transcription of maternal alleles of the zygotic genome, whereas, parental genomes contribute equally to the transcriptome of mature zygote[5]. Taken together, it is clear that maternal and paternal genome contribute equally to the transcriptomes of Arabidopsis embryos as early as zygotic stage. Genetic analysis of emb/+ mutants revealed that most emb/+ mutants displayed a typical Mendelian 3:1 segregation, supporting equal paternal and maternal contributions to the transcriptional activities of these EMB genes during early embryogenesis[57]. However, this result could not fully explain the model of paternal genome activation derived from the genetic studies. Del Toro-De Leon et al. demonstrated that among 49 emb mutants, wild-type paternal alleles of 40 EMB genes could not complement the phenotypes of respective emb embryo before 2 d after pollination[58]. This raises a striking question why paternal transcripts of EMB genes could be detected in early embryos, but could not exert their function.
It still remains to be elucidated how parents coordinate to regulate early embryogenesis, especially, regulate cell lineage specification. Comparative analysis of allele-specific transcriptome and morphological characteristics of apical and basal cell lineages of early proembryos demonstrated that parental contributions to the transcriptome is a different concept to the parental control of early embryogenesis. Clear evidence demonstrated that parental genomes contributed equally to the transcriptomes of both the apical and the basal cell lineages of early proembryos, but basal cell lineage development show a clear maternal effect[56]. The length of basal cell of hybrid proembryos is consistent with the maternal parents in the crosses between Arabidopsis Col-0 and Ler, and also between Wassileskija (Ws) and Ler. Besides basal cell length, suspensor cell length and cell number also show a maternal effect during basal cell lineage development[56]. Several potential mechanisms including specific roles of parent-of-origin genes, such as SSP, HOMEODOMAIN GLABROUS11/12 (HDG11/12), and maternal tissue-derived hormones, such as ovule integument-derived auxin, may be responsible for the parental effect on early embryo development. Relevant investigations have been discussed in recent reviews[17,21,38,59]. Although how maternal and paternal factors are integrated in early plant embryogenesis is still largely unknown, we can at least understand now that parental contribution to the transcriptomes and parental control of early embryogenesis are two different concepts. The transcriptome analysis is not sufficient to explain the specific parental roles in early embryogenesis.
Apical-basal embryo pattern formation
Establishment of apical domain of embryo
-
At the octant stage, the apical and basal domain of embryo proper can be easily distinguished from each other although both of them are derived from the apical cell. The upper tier of octant embryo will mainly develop into the shoot apical meristem (SAM) and cotyledons, whereas the basal tier of octant embryo will mainly divide to form the hypocotyl and embryonic root. The specification of two domains is marked by cell type specific molecular markers and regulated by different molecular pathways. At this stage, WOX2 is expressed in the upper tier cells, whereas WOX9 is activated in the basal tier of a octant embryo[35]. WOX2 and its three homologs WOX1, PRS and WOX5, act redundantly in regulating shoot patterning[39].
The morphology of SAM could be first outlined at the torpedo embryo stage. Establishment of SAM stem and organizer cells during embryogenesis is critical for postembryonic development[60]. Two well-known transcriptional factors, WUS and SHOOT MERISTEMLESS (STM), are critical for the establishment and maintenance of SAM. WUS mRNA was firstly detected in the upper inner cells of embryo at the dermatogen stage, and gradually became confined in the center of SAM[61]. WUS protein synthesized in the stem cell organizing center of SAM, and moved to the L1 and L2 layer cells in the central zone to activate the expression CLV3[62]. CLV3 that encodes a peptide hormone interacts with a LRR family receptor kinase CLV2 and a LRR receptor-like protein to in turn repress the expression of WUS through the formation of a feedback network[63,64]. STM, which encodes a KNOTTED-like homeodomain containing transcription factor, is essential for the initiation and maintenance the SAM together with WUS. STM mRNA was firstly detected in middle globular embryos, then in the apical domain of later globular embryos, and finally become restricted in the SAM between two cotyledons[65,66]. Strong mutation of STM led to lack of SAM[60].
In addition to WUS and STM, HD-ZIP II and III transcriptional factor family genes were reported to be involved in the establishment of SAM. HD-ZIP II transcriptional factor family genes including ARABIDOPSIS THALIANA3 (HAT3), ARABIDOPSIS THALIANA HOMEOBOX2 (ATHB2) and ATHB4 were also reported to be involved in SAM establishment. HAT3, ATHB2 and ATHB4 are expressed in early embryos and triple hat3 athb4 athb2 mutants lack an active SAM[67]. HD-ZIP III family genes PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ATHB8 and ATHB15 are expressed in early embryos, and involved in regulating shoot patterning[68−70]. REV, PHB, PHV, ATHB8 and ATHB15 play redundant roles in regulating the SAM establishment. rev phb double mutants show a shoot meristemless phenotype, whereas rev phv double mutants rarely show defects in SAM formation. The shoot meristemless phenotype of rev phb was enhanced by additional mutations in PHV and ATHB15, but not obviously enhanced by mutation in ATHB8, suggesting that REV and PHB play a major role in SAM establishment, whereas RHV and ATHB8 play a weak role in SAM formation[70]. HD-ZIP III transcription factors including PHB and REV are the targets of miR166/165[71]. ARGONAUTE 10 (AGO10), a central component of RNA-induced silencing complexes, could specifically interact with miR166/165 and promote miR165/6 degradation through the SMALL RNA DEGRADING NUCLEASE (SDN) pathway[72,73]. Accordingly, AGO10 is not only required for the WUS activity, but also for the maintenance of stem cells of SAM, potentially through WUS and miR165/6- HD-ZIP III two different pathways[72,74]. In addition to miR166/165, the surface cell layer produced miRNA394 is also involved in SAM formation through a non-cell autonomous manner[75].
Another important developmental event in establishing the apical domain of the embryo is the initiation of cotyledon primordia, which marks the shift from the radial symmetry to bilateral symmetry during embryogenesis. The sites of cotyledon primordia initiation show the accumulation of auxin indicated by the auxin response reporter DR5, suggesting the initiation of cotyledon primordia involves auxin dynamic[76,77]. The accumulation of auxin in the sites of cotyledon primordia formation is due to the polar auxin transport in the protodermal cells. Auxin polar transport in globular embryos was shown to be essential for the transition from radial to bilateral symmetry[77]. pin mutants including pin1, pin4 pin7 display severe defects in cotyledon formation[76]. Polar PIN1 localization is regulated by PINOID (PID) kinase through phosphorylation of conserved PIN motifs[78−80]. Consistent with the phenotype of pin mutants, no cotyledon phenotype was observed in pid wag1 wag2 triple mutants[81]. The expression patterns of CUP-SHAPED COTYLEDON1 (CUC1) and CUC2[82], two NAC family transcription factors required for cotyledon separation were found to be altered in pin1 mutants. AP2-type transcription factor DORNRÖSCHEN (DRN) and its homolog DRNL are direct targets of MP and also involved in cotyledon formation[83,84]. In addition to auxin related components, CLE19 is also reported to be involved in establishing the cotyledon. Although CLE19 is expressed in the cotyledon primordia, it is not required for cotyledon initiation, but contributes to the cotyledon growth[85].
Importantly, the cell fate of the apical embryo domain including SAM and cotyledon could be transformed into basal embryonic root fate[86]. In the topless-1 (tpl-1) mutant, the apical domain of an embryo developed into a root-like structure but not the shoot, resulting in the formation a double-root seedling without SAM and cotyledon. Correspondingly, topless embryos failed to express SAM marker gene STM and cotyledon marker gene KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 1 (KNAT1) , but expression of the root cell fate marker genes such as SCR[86]. TPL encodes a transcriptional corepressor and mutation in its putative corepressor, HISTONE DEACETYLASE19, leads to a similar defect in the apical domain, indicating that a transcriptional repression mechanism in the apical domain of embryo exists, which maintains shoot cell fate and prevent the embryonic root developmental pathway[87].
Establishment of hypophysis
-
The upmost cell in the basal cell lineage, also termed as hypophysis, will integrate into the embryo proper. During globular embryo development, the hypophysis will divide asymmetrically to generate two daughter cells with different developmental fates. The smaller lens-shaped cell will develop into the quiescent center, whereas the larger basal cell develops into the lower tier of stem cells. During past thirty years, great efforts have been made to investigate the mechanisms for hypophysis specification, which is closely linked to auxin signaling pathway and a set of transcriptional factors.
After asymmetric zygote division, PIN7 mediated basal-to-apical auxin polar transport is critical for apical cell lineage specification[52]. After the dermatogen embryo stage, the basal-to-apical auxin flow reverses to the apical-to-basal manner. The auxin flux reorientation is mediated by the redistribution of the auxin efflux regulator PIN1 and PIN7 in the apical and basal cell linages. In the apical cell lineage, non-polarly localized PIN1 began to localize to the basal side of the inner cells of the embryo proper to form a polar distribution. Whereas in the basal cell lineage, apically distributed PIN7 is re-localized to the basal side of the suspensor cells[52]. The re-localization of PIN1 and PIN7 lead to the accumulation of auxin in the hypophysis, as revealed by the auxin response reporter DR5. The accumulation of auxin is critical for hypophysis specification. Both auxin biosynthesis and transport mutants showed defects in hypophysis specification.
Besides critical roles of auxin biosynthesis and transport during hypophysis establishment, the auxin response is also essential for hypophysis specification. Auxin response in hypophysis is mediated by auxin response transcription factor MONOPTEROS (MP) and auxin response inhibitor BODENLOS (BDL), which affects hypophysis specification[88−90], which regulate hypophysis specification through a non-cell autonomous pathway. Several downstream molecular targets of MP have been identified in the process of hypophysis specification. The bHLH transcriptional factor TARGET OF MONOPTEROS7 (TMO7) is one target of MP, whose expression is regulated by MP in the provascular cells. TMO7 proteins could move into the hypophysis to regulate its asymmetric cell division[91]. In addition to MP, several auxin related factors including IAA10, AFR9 and ARF13 were also reported to be involved in hypophysis establishment[50,92].
Besides auxin related transcriptional factors, other transcriptional factors including zinc finger, AP2 and WOX family transcription factors were also reported in the specification of root meristem. The zinc finger transcription factors, NO TRANSMITTING TRACT (NTT), WIP DOMAIN PROTEIN4 (WIP4) and WIP5, were reported to regulate the initiation of the root meristem. NTT, WIP4 and WIP5 are expressed in the hypophysis and required for distal stem cell fate[93]. Besides NTT, WIP4 and WIP5, maternally expressed WIP1, WIP3 and WIP6 are also critical for cell fate specification in the embryonic root through a non-cell autonomous manner[94]. AP2 transcriptional factors, PLETHORA1 (PLT1) and PLT2, are essential for QC specification and maintaining root stem cell activities[95]. Consistent with their role in maintaining stem cell, PLT1 and PLT2 together with their two homologues PLT3 and PLT4 (also called BBM) are necessary for root formation[96]. Another important transcriptional factor in the hypophysis is WOX5, a member of WOX family genes. WOX5 is expressed in hypophysis and became restricted in the smaller lens-shaped cell after hypophysis asymmetric division[35]. WOX5 was reported to maintain the quiescent state of the cell by suppressing CYCLIN D activity[97]. WOX5 could also modulate the expression of the auxin biosynthetic genes to maintain the maximum auxin response and distal stem cell populations in the root tip[98]. In addition, WOX5 could also work as a mobile signal which moves from the QC to repress differentiation of columella stem cells through recruiting TPL/TPR co-repressors and the histone deacetylase HDA19 to silence CDF4 expression[99].
Radial embryo pattern formation
-
Radial pattern establishment is another important developmental event during early plant embryogenesis, which contains two major processes: the formation of epidermal layer, and primary differentiation of the ground and provascular tissue. In Arabidopsis, epidermal specification is initiated after first periclinal cell division of the octant embryo, which gives rise to the formation of an outer layer composed of eight protoderm cells, and an inner layer composed of eight inner cells. The outer protoderm cells divide anticlinal to extend the outer layer and will differentiate into the epidermis, whereas the inner eight cells divide longitudinally[100]. The four basal inner cells will develop into the ground and vascular tissue. The molecular mechanisms for epidermal specification are still largely unknown. The HD-ZIP IV transcription factor gene Arabidopsis thaliana Meristem Layer 1 (ATML1) is a protoderm-specific molecular marker. ATML1 is expressed in all embryonic cells from zygote and octant embryo stage, and then gradually become confined in the epidermal cells[101]. ATML1 cooperates with its homolog Protodermal Factor 2 (PDF2) to regulate epidermal specification. In atml1 pdf2 mutant, embryos are usually arrested at globular embryo stage without epidermal cell specification[102]. In addition to ATML1 and PDF2, several others genes including DEFECTIVE KERNEL1 (DEK1)[103] are also involved in epidermal specification.
As described above, periclinal cell division of octant embryo leads to compartmentation of protoderm and the inner tissues. The four basal inner cells will develop into the ground tissue and vascular tissues. Auxin-dependent transcription factor MP was shown to be critical for the initiation of ground tissue lineage during early embryogenesis[104]. For the ground tissue pattering and maintenance, GRAS family transcription factor SHORT-ROOT (SHR) and its target SCARECROW (SCR) were reported to be critical for the asymmetric cell division of the ground tissue cells. In shr and scr mutants, visible embryo development defects were observed at the heart embryo stage[105]. SHR and SCR could directly activate the expression of CYCD6;1 and thus regulate formative cell divisions[106]. Another transcriptional factor involved in ground tissue establishment is SCHIZORIZA (SCZ). In scz embryos, ground tissue stem cells show a distinct cell division pattern compared to that of the WT embryos, indicating that SCZ is critical for the establishment of ground tissue stem cells. Both mutations of SCR and SCZ resulted in the embryo lacking ground tissue[107]. In addition, CLE25 and its receptor kinases CLE‐RESISTANT RECEPTOR KINASE (CLERK) are expressed in the protophloem cell linage at the globular stage, which work together to regulate phloem development in Arabidopsis[108].
Vascular tissue establishment is also regulated by MP. Less cell files were found in the vascular tissue of mp embryos. This process is regulated by the basic helix-loop-helix (bHLH) transcription factor TMO5, another MP target. TMO5 appears firstly in the four vascular initial cells of globular embryos and is then restricted to the xylem precursor cells of heart-stage embryos[109]. TMO5 interacts with another bHLH transcriptional factor LONESOME HIGHWAY (LHW) to form a dimer to control the vascular tissue initiation during embryogenesis[109]. TMO5-LHW complex activates downstream LONELY GUY4 (LOG4) gene, which encodes an enzyme involved in biosynthesis of cytokinin and together with other LOG genes to control embryonic vascular tissue patterning[110]. The activities of the TMO5-LHW complex are antagonized by thermospermine synthase ACAULIS5 (ACL5), which promotes the translation of SAC51-LIKE (SACL) genes. SACL proteins could not only directly bind to the LHW, but also inhibit the activities of TMO5-LHW complex[111]. Interestingly, the expression of SACL genes is also regulated by TMO5-LHW complex[110], indicating that SACL proteins and TMO5-LHW form a feedback mechanism that controls vascular division. Recently, bZIP transcription factor GBF2 was shown to be able to interact with ARFs and regulate vascular gene expression in the process of vascular tissue specification[112], updating the gene regulatory network for vascular tissue identity.
Embryo maturation
-
Embryo development usually consists of two major phases: embryogenesis and embryo maturation[2,3]. As described above, basic embryo structures including apical-basal and radial patterns are established in the first phase. Embryo maturation is initiated around the heart embryo stage, and the switch from embryo morphogenesis to maturation involves dramatic changes in the storage components and gene expression profile. At the early heart stage, chlorophyll is accumulated in the protoderm, accompanied by proplastid maturation to the chloroplasts, and then an embryo turns to green and accumulates storage products including storage proteins and storage lipids[113]. The process of embryo maturation is tightly regulated by abscisic acid (ABA), which is initally produced in the maternal tissues, and then in the embryo itself. The ABA signal is tightly closed to the transcription program, and acts as a positive signal for dormancy during embryo maturation[114].
LEAFY COTYLEDON1 (LEC1), LEC2, ABSCISIC ACID INSENSITIVE3 (ABI3) and FUSCA3 (FUS3) are four master regulators in embryo maturation, and mutations in any of these four genes will lead to defects in embryo maturation. Although lec1, lec2, abi3 and fus3 show several common phenotypes including reduced seed storage protein gene expression[115] and decreased dormancy[3], they also display some distinct phenotypes such as chlorophyll accumulation, desiccation, and the accumulation of anthocyanins[115], suggesting that these four genes cooperate together to form a regulatory network for embryo maturation. LEC1 encodes a conserved heme-activated protein 3 (HAP3) subunit of the CCAAT box-binding transcription factor, whereas LEC2, ABI3 and FUS3 are B3 domain transcriptional factors. LEC1 is expressed in the embryo and endosperm, and is involved in both embryo morphogenesis and maturation. Briefly, LEC1 is essential for the accumulation of lipids and storage proteins, surviving desiccation. Expression of LEC1 in the endosperm, but not in the embryo itself, was found to be necessary for embryo maturation, suggesting that expression LEC1 in the endosperm could promote embryo development through a non-cell-autonomous pathway[116]. LEC1 and LEC2 could regulate the expression of ABI3 and FUS3. In addition, ABI3 and FUS3 could also regulate the expression of ABI3 and FUS3 themselves and each other, thus, forming a complex genetic network in embryo maturation[115,117].
Strategies for engineering apomixes and haploid induction
-
In addition to embryogenesis triggered by fertilization, embryogenesis could also be activated spontaneously in the absence of egg cell fertilization. This process is called apomixis, and naturally occurs in about 400 angiosperms. Although apomixes hardly occur in major crops and Arabidopsis, recent advances in plant reproduction make it possible to engineering apomixes and haploid induction in these species. In Arabidopsis, four major strategies have been used to induce the haploid formation (Fig. 3). The first is centromere-specific histone H3 (CENH3). cenh3-1 mutants display embryo lethal phenotype, which could be rescued by the expression of GFP-CENH3 and GFP-tailswap, a modified version of CENH3. cenh3-1 plants with the expression of GFP-CENH3 and GFP-tailswap could be used to induce haploids by crossing of the maternal and paternal lines[118]. The second approach is achieved through the use of the two sperm-specific proteins DMP8 and DMP9. DMP8 and DMP9 play a critical role in gamete fusion, especially in sperm-egg fusion[119,120]. Although exact mechanisms are still unknown, dmp8 dmp9 double mutant could be used as a paternal line to induce haploid plants when crossed with WT[121]. The third pathway is ectopic expression of BBM in the egg cell to induce haploid embryo development. In rice, expression of OsBBM in the rice egg cells could induce parthenogenesis[122], the ability of BBM in the initiation of embryo development was recently approved in Arabidopsis[123]. The expression BBM was not detected in the egg cells, but activated in zygotes shortly after fertilization[5,123]. Ectopic expression of Brassica napus BBM in the Arabidopsis egg cell could trigger parthenogenesis and lead to the formation of haploid plants at a low frequency[123]. Although the molecular mechanisms underlying embryo initiation in these three pathways may differ, they could be introduced into crops for breeding together with the MiMe (Mitosis instead of Meiosis) system, which turns meiosis into mitosis during reproduction[124]. Recently, a novel strategy was reported. Egg Cell Specific 1/2 are exclusively expressed in the egg cell and double mutant ecs1 ecs2 shows hemizygous phenotype and haploid offspring generation, therefore, the double mutant can be used as a maternal haploid inducing line[125].
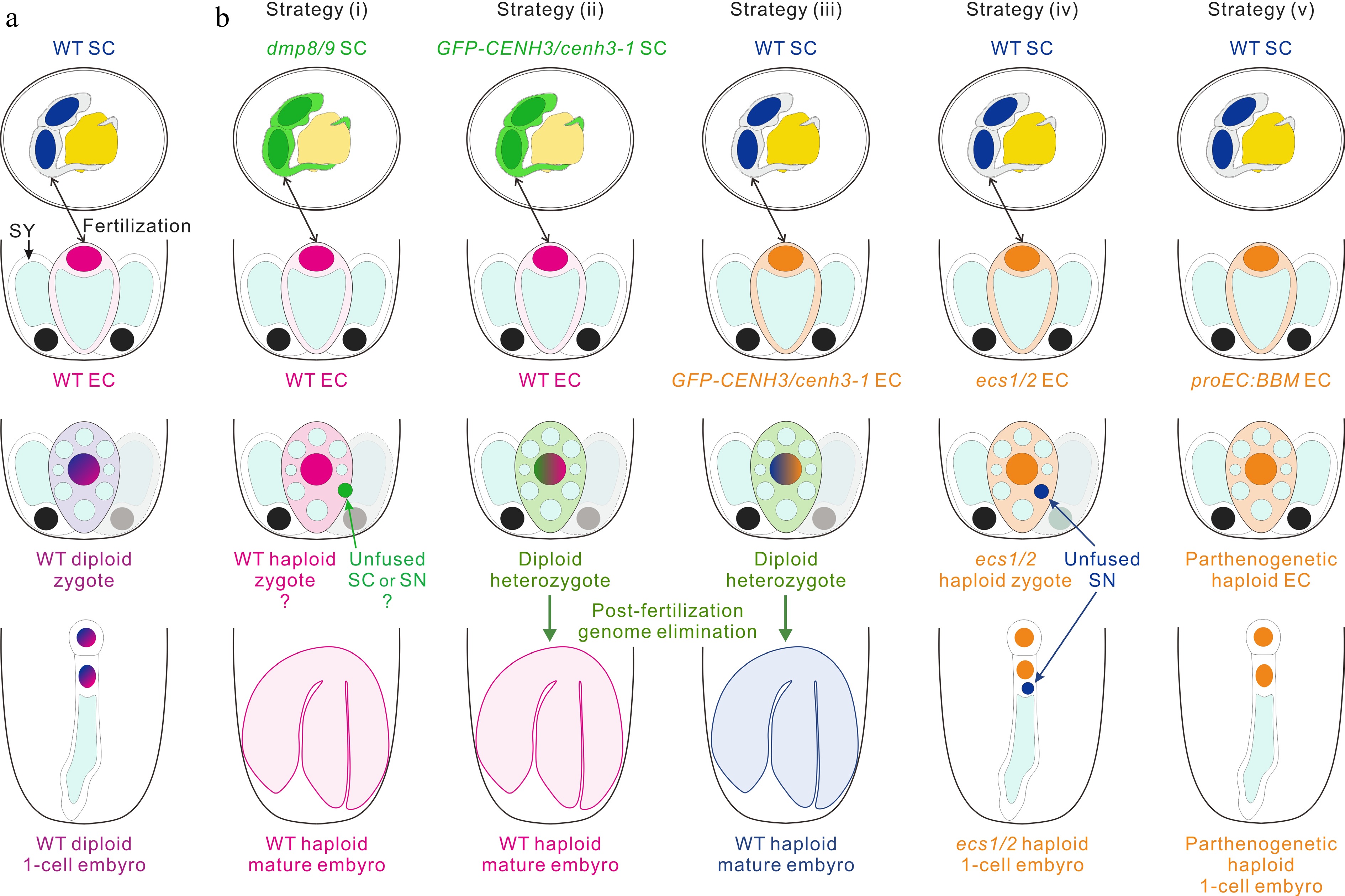
Figure 3.
Strategies for engineering haploid offspring. (a) The haploid sperm cell (SC) and egg cell (EC) fuse to produce a zygote, which then develops into a diploid embryo in wild-type (WT). (b) In vivo haploid embryo development. Strategy (i): the mutants of two sperm-specific genes DMP8 and DMP9 as paternal lines cross with the WT to induce haploid plants. Strategy (ii) and (iii): cenh3-1 plants with the expression of GFP-CENH3 or GFP-tailswap are used to induce haploids by cross of the paternal (ii) and maternal (iii) lines. Strategy (iv): the mutant of two egg cell-specific genes ECS1 and ECS2 as a maternal line and cross with the WT to induce haploid embryogenesis, which may be due to the karyogamy defect. Strategy (v): the egg cell-ectopic expression of Brassica napus BBM trigger parthenogenesis and lead to the formation of haploid plants. SY, synergid cell; SN, sperm cell nucleus.
-
During double fertilization, the sperm cell fuses with the central cell and the fusion product, primary endosperm cell, develops into the endosperm. The endosperm plays an important role in supporting embryo growth by supplying nutrients and other factors during seed development and germination[126,127]. In many monocots such as rice, wheat and corn, the endosperm persists until seed maturation and stores carbohydrates and proteins, which are the primary food source for human, whereas in dicots such as Arabidopsis thaliana, the endosperm is consumed by the embryo during subsequent seed development[126].
In Arabidopsis, right after fertilization, endosperm undertakes several rounds of mitosis without cell wall formation, resulting in syncytial endosperm. Then the syncytial endosperm initiates cellularization to produce cellular endosperm[128,129]. After cellularization, the endosperm experiences programmed cell death (PCD) and is believed to be gradually absorbed by the embryo, termed as endosperm breakdown (Fig. 1).
A recent report strongly suggests that endosperm development is an autonomously organized process in Arabidopsis, independent of embryogenesis[130]. In single-fertilization mutants dmp8 dmp9 and gex2, the central cell but not the egg cell is fertilized. In this case, endosperm initiation, syncytium formation, cellularization and PCD occur as in the wild type in terms of the cytological process and time course[130]. Here, we focus on the advances in the mechanism underlying endosperm development but not on the topic of the endosperm epigenetics regulation and endosperm effect on seed size, which have been previously well reviewed[126,131−133].
Initiation of primary endosperm nucleus division
-
Arabidopsis endosperm development begins with the division of the triploid primary endosperm nucleus, which initiates rapidly after the central cell fertilization and precedes the division of the zygote by several hours[134]. What molecular mechanism triggers the endosperm nucleus division remains largely unknown. The progress in recent years suggest two mechanisms, which negatively or positively control the initiation of primary endosperm nucleus division respectively.
Negative regulators of endosperm proliferation
-
Endosperm proliferation independent of fertilization is negatively controlled by Polycomb Group (PcG) proteins[135,136]. PcG proteins assemble in chromatin remodeling complexes and repress transcriptional activity of target genes[137]. The polycomb repressive complex 2 (PRC2) is responsible for trimethylated lysine 27 on histone H3 (H3K27me3), the hallmark of PcG-dependent gene silencing[138]. In Arabidopsis, PRC2 complex contains three FERTILIZATION INDEPENDENT SEED (FIS) genes, FIS1 or MEDEA (MEA), FIS2 and FERTILIZATION- INDEPENDENT ENDOSPERM (FIE)[139−143]. The fis mutants show autonomous division of the central cell, leading to endosperm-like development in the absence of fertilization[135,136]. The plant PRC2 complex also contains MULTICOPY SUPPRESSOR OF IRA1 (MSI1), a WD40 protein. Mutations in MSI1 cause a pleiotropic phenotype and causes defects in endosperm similar to those reported in fis mutants[144]. It was thus concluded that PRC2 negatively regulate endosperm growth and proliferation (Fig. 4a, b).

Figure 4.
Some regulatory pathways for the control of central cell and primary endosperm nucleus proliferation. (a), (b) The regulation network inhibiting central cell division before fertilization. PRC2 complex containing FIS1, FIS2, FIE and MSI1 inhibits central cell division and negatively regulate endosperm growth and proliferation. Dysfunction of FIS2 and FIE could activate auxin biosynthesis gene YUC10 and lead to autonomous division of central cell in the absence of fertilization. Similarly, adding the exogenous auxin 2,4-D or co-expressing auxin biosynthesis gene TAA1 and YUC6 in central cell also induce replication of the central cell. In addition, ectopically expression of CYCD3;1 or CYCD7;1 in central cell can induce central cell proliferation. (c) The endosperm proliferation is positively regulated by fertilization. Karyogamy facilitated by CDKA;1 is required for normal endosperm development. The sperm-derived miR159 inhibits its maternal targets, MYB33 and MYB65, in the central cell to promote the initiation of endosperm nuclear divisions. * indicates the ectopic genes expression in the central cell; ** indicates other components of the PRC2 complex.
It was suggested that the epigenetic regulators of the PRC2 complex block central cell division before fertilization by repressing the expression of auxin biosynthesis genes in the central cell[145]. Loss of FIS–PRC2 function in fis2 and fie could lead to ectopic expression of auxin biosynthesis gene YUCCA10 (YUC10) and auxin increases in autonomous endosperm. Auxin concentration is low in the central cell, while it increases in the endosperm[145]. Increased auxin by either adding synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) to unpollinated pistils or co-expressing auxin biosynthesis gene TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and YUC6 in the central cell is sufficient to induce replication of the central cell in the absence of fertilization (Fig. 4b). The activation of central cell division by the FIS-auxin module is partially dependent on the MADS-box transcription factor AGAMOUS-LIKE 62 (AGL62)[145]. AGL62 encodes a Type I MADS transcription factor and is expressed in the central cell and endosperm[145,146]. Loss of AGL62 function strongly suppressed autonomous endosperm development triggered by auxin or by fie in the ovule[144].
Retinoblastoma-related (RBR) inhibits the division of the central cell[147]. RBR genes belong to a conserved gene family in higher eukaryotes and are primarily known as negative regulators of the cell cycle[148,149]. RBR genes normally function to prevent inappropriate cell proliferation in animals. In Arabidopsis, a loss-of-function mutant of RBR1 results in nuclear proliferation in embryo sacs, including autonomous central cell proliferation because of a failure to exit the cell cycle[147]. D-type cyclins (CYCD) kinases can phosphorylate RBR to activate E2Fs[150], which promotes the transcription of genes required for the S phase[151]. Ectopic expression of CYCD3;1 or CYCD7;1 in the central cell leads to induced proliferation of the central cell[152,153] (Fig. 4b).
RBR1 also negatively regulates central cell division by regulation of transcription via chromatin-modifiers. RBR1 interacts with PRC2 member FIE and MSI1 and RBR1 are necessary for FIS2 expression in the central cell[154,155]. DNA METHYLTRANSFERASE 1 (MET1) is required for the maintenance of DNA methylation and heterochromatin and is critical for plant development[156,157]. MET1 maintains DNA methylation of FIS2 and inhibits its expression in pollen[158]. RBR1 binds to the promoter of MET1 to depress its expression, which may be important for FIS2 expression in the central cell[147] (Fig. 4a).
Positive regulators of endosperm proliferation
-
Besides the above negative control mechanism, endosperm proliferation was positively regulated by fertilization. Arabidopsis GCS1 (GENERATIVE CELL SPECIFIC 1) or HAPLESS2 (HAP2) is localized on the plasma membrane of sperm cells and is a critical fertilization factor in angiosperms[159,160]. When gcs1/hap2 sperm are released into the embryo sac, they attached to the egg and central cell but failed to fertilize them[161]. As a result, the central cell does not divide. The results indicate that sperm attachment without fusion cannot trigger division of the central cell[161].
A previous study showed that loss function of CYCLIN DEPENDENT KINASE A1 (CDKA;1) cause the production of a single sperm cell able to fertilize egg cell, while the unfertilized central cell could divide several times[162,163]. It had been proposed that a signal from the fertilized zygote triggers unfertilized central cell division[163]. However, other studies reported that the embryos produced by pollen carrying a single sperm do not trigger central cell division[164,165]. Later a re-analysis of the phenotype of cdka;1 showed that 32% of the cdka;1 pollen tube contained two sperm cells[166]. The sperm cell of cdka;1 could also fertilize the central cell, but the sperm nucleus did not decondense or fuse with the central cell nucleus (Fig. 4c). Nevertheless, sperm entry can trigger division of the central cell without karyogamy[166]. The result suggests that some factors from sperm cytoplasm promotes cell division in fertilized central cells.
Recently, Zhao et al. showed that microRNA159 (miR159) in sperm cytoplasm inhibits its maternal targets in the central cell to regulate the initiation of endosperm nuclear divisions[167]. miRNAs are 20–24 nucleotides small RNAs that play an essential role in various biological processes[168]. In Arabidopsis, miR159 includes three members, miR159a, miR159b and miR159c, which are sperm-enriched miRNAs[169,170]. The mir159abc triple mutant impairs the development of endosperm. The primary endosperm nucleus does not divide or just divide once or twice[167]. Two R2R3 MYB domain proteins, MYB33 and MYB65, are the targets of miR159[171,172] and are highly expressed in the central cell[167]. Both MYB33 and MYB65 are rapidly abolished in fertilized central cell, while loss of paternal miR159 leads to retention of MYB33 and MYB65 in fertilized central cell. Furthermore, overexpression of a miR159-resistant version of MYB33 in the endosperm inhibits endosperm nuclear division. The results suggest that MYB33 and MYB65 in the central cell inhibit the initiation of endosperm nuclear division, which can be abolished by miR159 in sperm cytoplasm upon fertilization (Fig. 4c). An earlier study showed that the female parts of the myb33 myb65 plant were fully fertile[172], implying that other proteins are of functional redundancy with MYB33 and MYB65 to inhibit the nuclear division in the central cell.
Syncytial endosperm
-
The morphogenetic events of the early stages of endosperm development in Arabidopsis were first investigated by Brown et al. [128] and later detailed by Li et al. and Boisnard-Lorig et al.[126,129]. The division of the primary endosperm nucleus occurs within 10 h after pollination and is much faster than the division of the zygote, which begins about 24 h after pollination. The first four nuclear divisions were synchronous and rapid with 3-6 h intervals[173]. In Arabidopsis, nuclear migration occurs in the direction from the micropylar to the chalazal chambers and nuclei are positioned in an equidistant manner[128,165]. The syncytial endosperm has three regions that become distinct with division: the embryo-surrounding region or micropylar endosperm (MCE), the peripheral endosperm (PEN) in the central chamber, and the chalazal endosperm (CZE)[128,129]. As the embryo sac expands after fertilization, the central vacuole enlarges and the cytoplasm of the endosperm syncytium assumes a peripheral position. At the globular embryo stage, the syncytial cytoplasm of the MCE surrounds the developing embryo, and the multinucleated PEN syncytium is a thin peripheral layer with evenly spaced nuclei.
The role of auxin in endosperm proliferation
-
Besides its role in promoting division of endosperm, auxin is necessary for proper proliferation of the endosperm. After fertilization, paternal YUC10 is expressed exclusively in the endosperm of developing seeds. As a result, the fluorescent signal of DII:VENUS, which is degraded when auxin presents, strongly decreased in the endosperm nuclei, indicating a postfertilization accumulation of auxin in the endosperm[145]. Mutants which are deficient either in auxin biosynthesis or auxin signaling have fewer endosperm nuclei with enlarged size than the wild type[145]. Ectopic expression of a non-canonical IAA, IAA32 in the endosperm under the control of the PHERES1 promoter impairs auxin signaling and causes auxin-deficiency phenotypes, thus leads to fewer endosperm nuclei[145]. How the auxin concentration control nuclear division in endosperm need further investigation.
The role of cyclins in endosperm proliferation
-
Cyclins were found to control the division of endosperm. B-type cyclins CYCB1;1, CYCB1;2 and CYCB1;3 are expressed during endosperm development[129,174,175]. The endosperm nuclei number in either cycb1;1 cycb1;2 or cycb1;2 cycb1;3 is much less than that of wild type 3 d after pollination (DAP)[175]. Consistent with this, overexpression of CYCD7;1 in the endosperm enhances the number of nuclei during syncytial endosperm development[153].
NAPHASE PROMOTING COMPLEX/CYCLOSOME subunit 11 (APC11) interacts directly with cyclin B1 to promote the degradation of cyclin B1[176]. Mutations of ZYGOTE-ARREST 1 (ZYG1), encoding the APC11, and two other APC subunits, APC1 and APC4, result in cell cycle synchronization defects in the endosperm[177]. Mutation in APC11 leads to cyclin B1 over-accumulation and transgenic expression of a non-degradable cyclin B1 in endosperms leads to unsynchronized mitosis in the syncytial endosperm[177], indicating that APC11 regulates the division of endosperm by control the level of cyclin B.
The genetic homeostasis influences endosperm proliferation
-
Some factors involved in modification of tRNA molecules, DNA replication and chromosome structural maintenance, were found to control the division of endosperm[178,179]. Most recently, it has shown that post-transcriptional modification of tRNA molecules played crucial roles in endosperm division[178]. AtTRM61/AtTRM6 complex methylate tRNAs to maintain the stability of initiator methionyl-tRNA (tRNAi Met)[180]. Mutations of AtTRM61/AtTRM6 resulted in endosperm nuclei cease at 5–8 free nuclei[178]. During the process of DNA replication, replication factor C (RFC) subunits are indispensable for S-phase checkpoint in yeast[181]. In Arabidopsis, AtRFC4 is indispensable for maintaining cell division of endosperm free nuclei[179]. Loss function of AtRFC4 resulted in defect endosperm with only six to eight free nuclei. Late S-phase entry and S-to-M transition were suppressed in rfc4 in root[179], it might be also the case in endosperm. The structural maintenance of chromosomes (SMC) gene family encodes chromosome scaffold proteins in yeast and plays a central role in chromosome segregation and condensation[182]. Mutations in SMC1, SMC2, and SMC3 resulted in defects of endosperm with less free nuclei showing huge volume, termed as a titan phenotype[183,184].
The role of microtubule in endosperm proliferation
-
Mutation in another two TITAN genes TTN1 and TTN5 also led to enlargement of endosperm nuclei with decreased number[185]. TTN5 encodes a ADP ribosylation factor (ARF) family of small GTP binding protein and probably functions in intracellular membrane trafficking or in cytoskeletal organization. TTN1 encodes a large regulatory protein known as tubulin-folding cofactor D (TFC D)[185].
The PILZ group of genes encodes proteins of the tubulin-folding complex, which is composed of five protein members including TTN1/TFC D[186]. Mutation in any of the five members led to the pilz phenotype, which is similar to titan[186]. TFC complex plays an important role in synthesis of α/β-tubulin heterodimers, the control of the ratio between α- and β-tubulin monomers and microtubule dynamics[187].
The augmin complex, consist of eight subunits including AUG1-8, plays an essential role in microtubule (MT)-dependent MT nucleation by recruiting the γ-tubulin complex to MT walls to generate new MTs[188]. ENDOSPERM DEFECTIVE1 (EDE1) is homologous to AUG8 and colocalizes with mitotic microtubules in vivo and binds microtubules in vitro[189]. EDE1 is essential for microtubule function and nuclear proliferation during endosperm development. At 6 DAP, ede1 endosperm displayed less than 10 enlarged nuclei, whereas the wildtype endosperm contained more than 100 normal nuclei at the same time[190]. The enlarged nuclei in ede1 endosperm might result from serious distortions in spindle MT remodeling during mitosis, since ede1-1 compromises the localization of augmin and γ-tubulin on the spindle and phragmoplast microtubules arrays[189].
Studies on ede1, titan, pilz mutants suggest that MT is important for the early nuclear division of endosperm. It is also supported by a recent study, which shows that treatment with oryzalin, an inhibitor of MT polymerization, results in overall MTs disruption and failed nuclear division in endosperm[173].
The role of small peptide signaling in syncytial endosperm development
-
Besides MT, small peptide signaling pathway also plays a role in syncytial endosperm development. CLE8, a member of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) family, is expressed in the embryo and surrounding endosperm region[191]. Mutation of CLE8 results in defective early nuclear migration and disorganization of the nuclear endosperm[191]. CLE19, another member of CLE family mainly expression in epidermal cells of heart-stage embryo, is also shown to affect Arabidopsis endosperm development[85]. Disturbing CLE19 function results in endosperm showing reduced nuclear number with big nuclei[85]. How CLE8 and CLE19 regulate syncytial endosperm development needs further investigation.
Cellularization of endosperm
-
At the globular stage of embryos (5 DAP), the formation of cell walls in the syncytial endosperm is initiated in the MCE[129,173]. This process, termed as cellularization, was reported to proceed as a wave across the PEN towards the CZE[128]. Plant cytokinesis requires de novo secretory trafficking[192]. The cell wall formation is mediated by specialized types of phragmoplasts and cell plates[193,194]. Genetic analysis suggests that endosperm cellularization and somatic cytokinesis share many components[195]. During endosperm cellularization phragmoplasts are observed at the junctions of opposing ends of radial microtubules that encircle each of nuclei[128]. The timing of endosperm cellularization is critical for seed size and embryo development[196,197]. Generally, precocious endosperm cellularization leads to small seeds, while delayed endosperm cellularization results in larger or aborted seeds[146,196]. The mechanisms controlling endosperm cellularization and their timing have been the research focus in past decades.
Endosperm cellularization needs proper vesicle trafficking and phragmoplasts assembly
-
ARABIDOPSIS FORMIN HOMOLOGUE 5 (AtFH5) encoding a member of the formin family that is a potent actin nucleator induces the assembly of actin filaments[198]. AtFH5 is localized in the vesicle and targeted to the developing cell plate[199,200]. Loss function of AtFH5 shows a delayed cytokinesis in the endosperm but not in vegetative tissues[199]. This may result from the defect of assembly of actin filaments, which is critical for vesicle trafficking[200]. The KNOLLE gene, encoding a member of the syntaxin family required for docking and fusion of vesicles at the target membrane, plays an important role in cytokinesis[201]. KNOLLE is located in the cellularizing endosperm and is critical for endosperm cellularization since most knolle endosperm are not cellularized[195,202]. The HINKEL gene encodes a kinesin-like protein involved in reorganization of phragmoplast microtubules and mutation of HINKEL interrupts endosperm cellularization[203]. It has been reported that mutant for SPÄTZLE, the gene product has not yet been identified, affects cellularization of the endosperm but not the embryo[195], indicating some factors specifically function in endosperm cellularization.
The role of AGL62 in endosperm cellularization
-
AGL62 is a major negative regulator of endosperm cellularization[146]. AGL62 expression is strong during the syncytial phase and then declines abruptly just before cellularization[146]. Based on the fact that the endosperm is prematurely cellularized in agl62, it has been suggested that AGL62 is required for suppression of cellularization during the syncytial phase[146]. AGL62 also play a role in interploidy crosses by regulating endosperm cellularization[204]. Interploidy crosses between diploid maternal and tetraploid paternal plants (2n × 4n) cause increased expression of AGL62, which correlates with endosperm cellularization failure and seed abortion[204]. Whereas crosses of tetraploid maternal and diploid paternal plants (4n × 2n) cause precocious endosperm cellularization with decreased expression of AGL62[205]. It can be concluded that AGL62 is a dosage-sensitive and negative regulator of endosperm cellularization.
AGL62 is substantially marked by H3K27me3 in the endosperm and its expression is under negative control of the FIS-PRC2 complex[197]. Loss of FIS-PRC2 function causes prolonged expression of AGL62 and failure of endosperm cellularization[197]. Maternal loss of AGL62 can partially restore endosperm cellularization in fis2 seeds[197].
AGL62 plays a negative regulatory role for endosperm cellularization by activating auxin biosynthesis[145]. In agl62 endosperm, it has a significant reduction of auxin accumulation and auxin biosynthesis gene expression[145]. Increased auxin by adding synthetic auxin (2,4-dichlorophenoxyacetic acid; 2,4-D) can partially rescue the endosperm defect of agl62 seeds[145]. How AGL62 regulates auxin biosynthesis in Arabidopsis remains unknown. It has been shown that FveAGL62 can activate the expression of FveYUC10 by inhibiting the transcription of FveATHB29b and FveATHB30, which are transcription factors and act to repress auxin biosynthesis[206]. Whether similar pathways also function in Arabidopsis needs further investigation.
The role of IKUs in endosperm cellularization
-
A group of mutants show haiku (iku) phenotype with precocious cellularization of the endosperm[196,207]. These mutants, including iku1, iku2 and miniseed3 (mini3), have small but viable seeds[196,207]. IKU1 encodes a VQ domain protein of unknown function[208]. IKU2 encodes a leucine-rich repeat transmembrane kinase and MINI3 encodes a WRKY10 transcription factor[207]. An iku-like phenotype has been observed in the mutant short hypocotyl under blue 1 (shb1)[209]. Genetic analysis shows that the four genes are in the same pathway[196,207,209]. IKU1, IKU2 and MINI3 are expressed in the syncytial endosperm after fertilization[207,208]. The expression of IKU2 depends on MINI3[207]. It has been suggested that IKU1 and MINI3 form a complex to regulate the expression of IKU2[207]. SHB1 acts on upstream of MINI3 and IKU2[209]. MINI3 recruits SHB1 to its own and IKU2 promoters to activate the transcription of MINI3 and IKU2[210]. MPK10, a member of mitogen-activated protein kinase (MAPK) family, interacts with MINI3/WRKY10 and inhibits the transcriptional activity of WRKY10 to negatively regulate the seed size[211].
IKU2 gene is under epigenetic control of the FIS–PRC2 complex to regulate duration of endosperm proliferation[212]. H3K27me3 marks are enriched over the AtIKU2 locus at 4 to 5 DAP, resulting in the silencing of IKU2 and causing inhibition of the endosperm proliferation. AtMEA catalyzes the trimethylation of histone 3 lysine 27, as IKU2 expression is increased in mea/mea. Interestingly, the expression level of IKU2 might drive the extent of endosperm persistence in different species. In Brachypodium and rice, which endosperms continue to proliferate until the maturity of the seeds, endogenous IKU2 showed persistent expression up to 20 DAP. Correspondingly, no H3K27me3 marks were enriched at the BdIKU2 and OsIKU2 locus[212].
The role of plant hormones in endosperm cellularization
-
ABSCISIC ACID (ABA) signal acts upstream of SHB1-IKU pathway to negatively control endosperm cellularization[213]. Endosperm cellularization is delayed in mutants of ABSCISIC ACID DEFICIENT2 (ABA2), which functions in ABA biosynthesis[213,214]. Functional loss of ABA INSENSITIVE 5 (ABI5), a transcription factor critical for ABA signaling transduction, resulted in seed size increase[213]. SHB1 RNA accumulation was significantly upregulated in the aba2 or abi5 and was downregulated by the application of exogenous ABA or overexpression of ABI5[213]. ABI5 directly binds to the promoter of SHB1 to negatively regulate SHB1[213]. TERMINAL FLOWER1 (TFL1) plays an essential role in endosperm cellularization by interacts with and stabilizes ABI5[215]. Interestingly, TFL1 is a mobile protein which generates in the chalazal endosperm and is transported to the syncytial peripheral endosperm[215]. The trafficking of TFL1 is mediated by ras-related nuclear GTPases, which also functions in endosperm cellularization[128,216].
In contrast to ABA, another plant hormone Brassinosteroids (BRs) acts upstream of SHB1-IKU pathway to positively control endosperm cellularization[217]. BR-deficient and -insensitive mutants have small seeds. The weak BR-deficient mutant de-etiolated2 (det2) showing delayed endosperm cellularization[217]. BRs modulate endosperm by directly binding to the promoter of SHB1, IKU1, IKU2 through BRASSINAZOLE-RESISTANT 1 (BZR1) to regulate the transcriptional levels of SHB1, IKU1, IKU2 and MINI3[217]. In addition, genetic analysis suggests BR inhibits the expression of APETALA 2 (AP2) to positively control endosperm cellularization[217]. AP2 is a negative regulator of endosperm cellularization since endosperm cellularization of the ap2 mutant occurs later than that of wild-type[218].
In addition, Cytokinin (CK) signal act downstream of SHB1-IKU pathway to control seed size[219], possibly through regulating the timing of endosperm cellularization. Cytokinin Oxidase 2 (CKX2) is involved in catabolism of cytokinin. The expression of CKX2 is directly activated by MINI3/WRKY10 and overexpressing CKX2 can partially rescue iku2 phenotypes[219].
Endosperm breakdown
-
In Arabidopsis, after cellularization, the endosperm is gradually degraded and only the peripheral endosperm layer remains intact and alive at maturity stage[128]. During endosperm breakdown, the endosperm experiences PCD which appears in endosperm cells at 5 DAP and becomes widespread at 6 DAP[130].
The mechanism underlying endosperm breakdown remains very poorly understood until the discovery of an endosperm specific bHLH transcription factor ZHOUPI (ZOU), also known as RETARDED GROWTH OF EMBRYO1 (RGE1)[220,221]. ZOU is expressed specifically in early endosperm, subsequently being confined to the region of endosperm surrounding the embryo (ESR)[220]. Loss function of ZOU shows defect in endosperm breakdown, which restricts embryo expansion[220−222]. The ZOU protein is widely conserved in plants and heterologous expression of ZOU orthologs in rice, maize and soybean can partially complement the seed phenotypes of zou[220,223−225], suggesting that ZOU plays a evolutionarily conservative function in endosperm breakdown.
Loss function of another bHLH-encoding gene INDUCER OF CBP EXPRESSION 1 (ICE1) causes a zou-like phenotype[226]. ZOU and ICE1 may form homodimers to regulate the expression of target genes in the developing endosperm[226]. It is proposed that ZOU indirectly triggers PCD of the endosperm by regulating the expression of cell wall-modifying enzymes, thus to weaken endosperm cell walls and allow the growing embryo to exert mechanical pressure on the endosperm cells[227]. However, Xiong et al. showed that PCD of endosperm is an autono/>mously process independent of embryo[130], suggesting that mechanical pressure from developing embryo is not the major cause of the PCD in endosperm.
Proteases have also been implicated in the regulation of many plant PCD processes and some protease genes have been found to be expressed in association with endosperm degeneration[228]. ZOU regulates the expression of several proteases, including ABNORMAL LEAF SHAPE1 (ALE1), BIFUNCTIONAL NUCLEASE1 (BFN1) and the PCD-associated PUTATIVE ASPARTIC PROTEINASE A3 (PASPA3)[220,227]. ALE1 is involved in the interaction of endosperm-embryo but not in endosperm breakdown[220,229], whereas whether PASPA3 and BFN1 are functional in endosperm breakdown remains unknown. Thus, how ZOU triggers PCD of the endosperm also needs further investigation.
-
In seed plants, the integument and nucellus envelope the embryo sac to make up the outer layer of the ovule structure[230]. The number of integuments in an ovule varies between different plant species, but most angiosperms own the bitegmic ovules which contain both inner and outer integuments composed of multiple cell layers. The integuments connect with the vascular tissues of maternal funiculus at chalazal region to ensure the nutritional supply for ovule development. The micropyle is constructed by the integuments for the pollen tube entering embryo sac. After fertilization, the integuments develop into a seed coat to enclose the embryo and its surrounding nutrient tissue[231]. For flowering plant species, the nutritive tissue in seeds can be either the fertilized central cell-originated endosperm or the maternal nucellus-derived perisperm. Alternatively, the reserves can also be transported to the cotyledon in Arabidopsis, resulting in only one layer of endosperm cells which is tightly attached with seed coat wrapping the embryo in mature seeds[231].
The seed coat is a protective outer envelope surrounding the embryo to protect it from the external damages including mechanical injury, ultraviolet irradiation, toxic chemicals and pathogen infection etc. The impermeable barrier providing by seed coat restricts the exchange of water and air between the outside environment and the seed to maintaining the coat-imposed embryo dormancy. When the favorable germination conditions and the most proper period for seedling formation exist, seed coat, in turn, offers the means for the initial water uptake. Thus, the seed coat plays multiple roles in seed dormancy and germination[232].
Here, using Arabidopsis as a model, we first introduce the anatomy of seed coat and then focus on describing the molecular network that regulates the post-fertilization development of the endothelial layer (the innermost layer of inner integument (ii)) and the epidermal layer (the outermost layer of outer integument (oi)), especially their cell specification.
The morphology of seed coat
-
In Arabidopsis, the three layers of cells in the inner integument and the two layers in the outer integument, which are named as ii1, ii1', ii2, oi1 and oi2 respectively, undergo a dramatic growth within a short period after double fertilization, rapidly cell dividing and expanding, and eventually adopting distinct cell fates[231−234] (Fig. 1).
The post-fertilization development of inner integument
-
The innermost integumentary cell layer (ii1), namely endothelium, synthesizes the flavonoid compound proanthocyanidins (PAs, also known as condensed tannins) and accumulates them in the central vacuole. Following the seed desiccates, the originally colorless PA oxidizes and forms brown pigments which are apparently released from the endothelial cells and impregnate the inner three cell layers to impart the color for testa in mature seeds[235]. In addition, the cell wall of endothelial cells on the side facing the endosperm deposits a cutin-based apoplastic barrier to separate the maternal and zygotic tissues[236,237]. By contrast, the parenchymatic ii1' and ii2 layers, the two other ii layers, do not undergo further cell differentiation, and are the first of the five integumentary cell layers to activate PCD. During this PCD process, cell death occurs in the ii1' layer followed by cell death in the ii2 layer, which leads to the two ii layers to eventually collapse in mature seeds. A caspase-like cysteine proteinase δ Vacuolar Processing Enzyme (δVPE) is specifically and transiently expressed in ii1' and ii2 layers at the early stage of seed development, and regulates the PCD in these limited cell layers to ensure proper seed coat formation[235,238]. In addition, the outer integuments express NACREGULATED SEED MORPHOLOGY1 (NARS1) and NARS2 (known as NAC2 and NAM, respectively), which may be also involved in PCD in the inner integument[239].
The post-fertilization development of outer integument
-
At the beginning of seed coat development, the cells of both outer integument layers have a large single vacuole and produce the starch-containing amyloplasts before diverging in cell fate. The starch granules in the oi1 layer (subepidermal layer) distribute in the inner periclinal plane of the cells facing the embryo, while the ones in the oi2 layer (epidermal layer) are characteristically accumulated at the center of the outer periclinal plane of the cells staying away from the embryo. Following vacuole division and starch degradation, the cells of the oi1 layer gradually atrophy, and eventually, only a thick secondary cell wall attached to the oi2 layer is left, forming a palisade layer[233]. Meanwhile, the cells of the oi2 layer synthesize and secrete mucilage, a pectinaceous carbohydrate, to the apoplastic space between the outer primary wall and the protoplast. With mucilage deposition and the central vacuole shrinking, the protoplast containing starch grains are forced into the center of the oi2 cells and form the cytoplasmic column. As the vacuole disappears and starch grains disintegrate, the newly created secondary cell wall occupies the space of the cytoplasmic column to establish the columella. Finally, the epidermal cells appear as a columella surrounded by the doughnut-shaped desiccative mucilage[232,233,240]. When the seed coat matures, all cells of five integumentary layers are lifeless (Fig. 1). Prior to this, ii1 and oi2 layers undergo more complex cell differentiation and substance synthesis than the other ii1', ii2 and oi1 layers, suggesting that they may need more elaborate regulatory mechanisms.
The regulation of endothelial development
-
The endothelial cells are characterized by the production of PAs, which increase the thickness and mechanical strength of the seed coat to prevent the the seed from being crushed easily, and modify the permeability properties of the seed coat to reinforce coat-imposed dormancy[241]. Thus, by screening the Arabidopsis mutants with distinguishable defective phenotypes such as the altered PAs level, the abnormal seed coat pigmentation, and the reduced seed dormancy etc, a series of genes regulating the development of the PA-accumulating endothelial cells were identified and then cloned[242,243] for our comprehensive understanding of cell differentiation and functional specialization in the ii1 layer.
Key regulators involved in endothelial patterning
-
TRANSPARENT TESTA 16 (TT16) / ARABIDOPSIS BSISTER (ABS) encoding the MADS domain protein is considered to be a global regulator lying upstream of the other transcription factors, controlling the endothelial cell specification[232,244]. Deletion of TT16 functions does not affect the ovule development and the inner integument integrity before fertilization. However, at the two-celled pro-embryo stage, in contrast to wild-type (WT) endothelial cells, which have a dense cytoplasm, rectangular shape and have an orderly arrangement, the tt16 endothelial cells exhibit a modified irregular morphology and vacuolization, indicating that TT16 is responsible for regulating endothelial cell architecture prior to pigment deposition. At the subsequent globular and heart-embryo stages, the altered shape of the tt16 endothelium is particularly obvious and no longer distinguishable from two ii1' and ii2 parenchymal layers[244]. In addition, TT16 also regulates the proximal-distal developmental patterning of the ii1' layer[245]. The ii1 layer undergoes a periclinal cell division to generate the ii1' layer at the end of ovule development[230,235]. TT16 mutation leads to the occurrence of an unwanted periclinal division of the ii1' layer that gives rise to an extra inner integument layer (ii'') in the micropylar zone, meanwhile, the morphology of the original ii1' and ii2 layers is disturbed[244,245]. Remarkably, the outer integument develops normally in tt16; therefore, TT16 regulates specifically the cell differentiation and to some extent the integrity of whole inner integument, particularly ensures the endothelium cell identity after fertilization[244].
Another key transcription factor that regulates endothelium differentiation is the WIP zinc-finger protein TT1. TT1 expresses specifically in the developing ovules and young seeds. tt1 mutant seeds have the endothelial cells with completely lack granules, which is similar to the defect phenotypes in tt16[234,246]. Further analyses reveal that TT1 modulates the polarity of endothelium cell expansion along proximal-distal axis. Compared to WT, the tt1 endothelium cells appear an increased cell area; moreover, their morphology showed more elongation in the chalazal region and rounder in the micropylar region. Besides, TT1 also regulates the cell expansion of the ii1' layer[234]. TT1 is regulated by TT16 at the chalazal region, thus a TT16-TT1 transcriptional module may contribute to establish chalazal endothelium patterning[234].
Major genes that regulate PA metabolism
-
A class of important structural genes participating in PA metabolism encodes a series of catalytic enzymes. The initial synthetic steps of different flavonoid sub-pathways are common, namely, chalcone synthase (CHS/TT4) catalyzes 4-coumaroyl-CoA and 3×malonyl-CoA to produce naringenin chalcone which is converted as naringenin by chalcone isomerase (CHI/TT5); later, hydroxylation of naringenin is preformed by flavanone 3-hydroxylase (F3H/TT6) and flavonoid 3'-hydroxylase (F3'H/TT7) to synthesize dihydroflavonol. Then, an NADPH-dependent dihydroflavonol reductase (DFR/TT3) leads to the production of leucoanthocyanidin (flavan-3,4-diol), which is the last common intermediate in the anthocyanin and proanthocyanidin biosynthesis[243,247]. Leucoanthocyanidin is converted to catechin by leucoanthocyanidin reductase (LAR) encoded by BANYULS (BAN) gene. Finally, Leucoanthocyanidin and catechin (PA precursors) are transported to vacuoles where they are polymerized to form PA. Also, because Leucoanthocyanidin is able to be catalyzed by leucocyanidin dioxygenase (LDOX/TT18/TDS4) to eventually form anthocyanin when BAN is dysfunctional, PA precursors are deficient and anthocyanin, instead of PA, is accumulated in the ban endothelial cells[247−251].
Another class of proteins regulating PA biosynthesis involves in the transport and sequestration of some PA precursors from their synthesis site in cytosol to the specific cellular compartments. In Arabidopsis, an H+-ATPase, a putative Multidrug and Toxic Compound Extrusion (MATE) secondary transporter, and a glutathione-S-transferase (GST) encoded by AHA10, TT12, and TT19 respectively are thought to manage the vacuolar transport of the PA precursors[251−255]. All aha10, tt12 and tt19 mutants exhibit an abnormal vacuolar biogenesis and PA depositional patterning in the endothelial cells of seed coat[252−254].
Transcriptional control of PA biosynthesis
-
Both TT16 and TT1 not only control the morphogenesis and architecture of the endothelium, but are also required for PA biosynthesis. TT16 triggers BAN expression to facilitate PA accumulation in the seed body area of the endothelium. However, it's worth noting that tt16 and tt1 mutation do not affect PA accumulation in the chalazal and micropylar area of seeds, suggesting that other regulators may be involved in the PA synthesis process in these two regions[244,246].
TRANSPARENT TESTA GLABRA (TTG1), a WD40 repeat (WDR) protein, often forms complexes with different MYB and bHLH transcription factors to specify the differentiation of certain cell types in plants[256−258]. During endothelial cell development, TTG1 synergizes with a bHLH transcription factor TT8 (bHLH042) and an endothelium-specific MYB transcription factor TT2 (MYB123) to establish a specific stable ternary transcription complex which activates the expression of the flavonoid biosynthetic related target genes and regulates the PA biosynthesis[247,259,260]. The normal expression of the PA late biosynthetic genes (LBGs), DFR/ TT3, BAN and TT12, depends on TT2 cooperating with TTG1 and TT8, however, the transcriptional activation of the PA early biosynthetic genes (EBGs) (CHS/TT4, CHI/ TT5, F3H/TT6 and F3'H/TT7) is independent of TT2. Any other related Arabidopsis MYB proteins cannot replace TT2 to activate BAN; and the ectopic TT2 expression is sufficient to trigger BAN in some vegetative tissues where the TTG1 existence, indicating that the endothelial cells-specific TT2 specifically plays key role in the genetic control of the flavonoid late metabolism and PA synthesis during the endothelium development of immature seeds[247,259,260]. PA is completely absent in ttg1-1, tt2-3, and tt8 seeds, indicating that the TTG1-TT2-TT8 complex controls PA synthesis throughout the seeds[243,247,261].
In addition, the TTG1-TT2-TT8 complex also activates the expression of TTG2 (WRKY44), a plant-specific WRKY transcription factor, to control the PA production[261]. But TTG2 is not necessary for BAN transcription, suggesting that it may regulate PA production through other pathways[235]. The subsequent studies showed that TT8 and TTG2 may form an unpredicted positive feedback regulatory loop[262]. Furthermore, TTG2 binds to the upstream regulatory region of TT12, and the ectopic expression of TT12 can partially rescue the pigmentation defects in ttg2-1 seed[261,263]. The regulation of TTG2 on TT12 depends on the interaction between TTG2 and TTG1, implying that TTG2 can be integrated into the TTG1-TT2-TT8 complex or form a new separated transcriptional complex with TTG1 (TTG1-TTG2 complex)[263].
The regulation of epidermal development
-
The epidermal cell, also called the mucilage secretory cell of theseed coat, is characterized by synthesizing mucilage and establishing columella[233]. Mucilage is an extracellular matrix which is hypothesized to own multiple functions including affecting seed dispersal, preventing seed from the digestion in animal's gut, governing seed germination, maintaining seed hydration and seedling establishment etc[264,265]. The mechanisms regulating the synthesis, modification, secretion and stabilization of mucilage have been systematically discussed in previous reviews. In this section, we mainly focus on the cell differentiation and morphological establishment of seed coat epidermis (oi2 layer) in Arabidopsis, including epidermal cell identity, transcriptional regulation of mucilage biosynthesis, and secondary cell wall (SCW) formation.
Key regulators involved in epidermal differentiation
-
The Arabidopsis epidermal cell differentiation has been divided into five stages: (1) fertilization-triggered cell growth of outer integument; (2) amyloplast accumulation and cytoplasmic rearrangement initiation; (3) mucilage production and cytoplasmic column formation; (4) SCW production; (5) desiccation[264]. APETALA 2 (AP2) is a vital regulator of epidermal cell specification which appears to be required for epidermis differentiation to move beyond early stage 2 and affects extensively epidermal cell features, thus it could be the outer integument counterpart of TT16 in defining cell type[266]. In abnormal ap2-6 seeds, two cell layers of outer integument fail to differentiate into the epidermal and palisade layers after initial enlarging, vacuolation, and amyloplast accumulation. Finally, only a few crushed cells surround the pigmented endothelium layer, while mucilage production and secondary wall synthesis are absent in mature ap2-6 seeds[266]. Similarly, the outer integument-expressed NARS1/NAC2 and NARS2/NAM also take part in the epidermis differentiation. The cells of oi1 and oi2 layers do not continue to differentiate after undergoing the early development, and consequently lose mucilage and columella in nars1 nars2 seeds[239].
A TTG1 regulatory complex, analogous to the TTG1-TT2-TT8 ternary complex in governing endothelium pattern formation, regulates the epidermal differentiation and mucilage biosynthesis of seed coat. TT8 together with a partially redundant bHLH transcription factor ENHANCER OF GLABRA 3 (EGL3) interacts with TTG1 to promote mucilage production. Moreover, MYB5 show partial function redundancy with TT2. Therefore, in the epidermal TTG1 complex, EGL3 and TT8 serve as bHLH transcription factors, and MYB5 and TT2 act as MYB transcription factors to impact epidermal differentiation[267,268]. The phenotypic observations of ttg1, egl3 tt8 and myb5 tt2 mutants reveal that the absence of any component of this MYB-bHLH-WD40 transcriptional complex results in the abortive epidermal differentiation accompanied by a loss of mucilage production and aberrant columella shape[266−270].
The TTG1-TT8/EGL3-TT2/MYB5 complex directly triggers the expression of the downstream genes encoding WRKY transcription factor TTG2 and homeodomain transcription factor GLABRA2 (GL2). In turn, GL2 physically interacts with trihelix transcription factor DE1 BINDING FACTOR 1 (DF1), and then both proteins activate synergistically a mucilage biosynthesis gene MUCILAGE MODIFIED 4 (MUM4) to produce mucilage[261,270,271]. Thus, the mature seeds of ttg2, gl2, df1 and mum4 with aberrant epidermal cell morphology produce less mucilage than WT, the holistic symptom similar to but less severe than that of ttg1[261,266,270,271]. Moreover, the epidermal differentiation is also controlled by MYB61, and its loss of function phenotype in epidermis is similar with ttg2, gl2, and df1. However, MYB61 deletion can aggravate the epidermal abnormal phenotype of ttg1. MYB61 is not able to modulate MUM4, suggesting that MYB61 may function in a distinct genetic pathway from that of the TTG1-mediated regulator network[270,272].
Developmental regulation of epidermal SCW
-
The SCW development of epidermal cells covers the thickening of the radial cell wall (RCW) and the formation of a polarized columella. Several genes encoding cellulose synthase subunits act on the dynamic process of the SCW synthesis and deposition in RCW. CELLULOSE SYNTHASE9 (CESA9) expresses preferentially in seeds, and its dysfunction can characteristically reduce the cellulose content in seeds which leads to a syndrome of the abnormal epidermal cell morphogenesis including depleted RCW, varied cell size and cell shape, altered internal angle uniformity, and defective barrier function of the seed coat[273]. Similar to CESA9, CESA2 is very important for the RCW reinforcement and correct epidermal cell shape, but not fundamental for the cellulose synthesis in other cells[273,274]. Unlike CESA9 and CESA2, CESA5 not only serves in RCW integrality and cell shape uniformity of epidermal cells, but also functions in the mucilage stabilization through the production of rays of cellulose microfibrils[274]. Besides, crystalline cellulose deposition into the RCW of cobra-like2 (cobl2) and quasimodo2 (qua2) mutants suggests that in addition to cellulose synthase, a glycosylphosphati- dylinositol-anchored COBRA-LIKE protein COBL2 and a pectin methyltransferase QUA2 also control the RCW reinforcement of epidermal cells[275,276]. In contrast to five genes mentioned above, both MYB75 and KNAT7 negatively regulate RCW thickness through unrevealed transcription networks. The deletion of these genes results in a thicker RCW[277].
The directed deposition of mucilage into the apoplast together with the large vacuole shrinking in epidermal cells induces the formation of a central cytoplasmic column, wherein SCW materials are gradually accumulated to build the columella[233]. CESA2, CESA9 and CESA5 play redundant roles in the columella synthesis[274]. The arabinogalactan protein SALT-OVERLY SENSITIVE5 (SOS5) that mediates mucilage adherence and organization works with CESA5 is also required for normal columella formation in a synergistic manner. The cesa5-1 sos5-2 columella often appear much smaller than that in WT and sometimes completely absent. This severe columella defects seem to originate from an altered morphology of the cytoplasmic column caused by the reduced mucilage adherent properties and enlarged mucilage pocket structure[278]. Besides, some other examples also support the hypothesis that mucilage is capable of determining the morphology of the cytoplasmic column, and subsequently, the shape of the columella. In contrast to cesa5 sos5, the less mucilage in ttg1-1, ttg1-1 myb61-1, myb5-1, myb5-1 myb23, gl2-1, df1-1, df1-1 gl2-8, mum4-1 and df1-1 mum4-2 mutants results in a smaller mucilage pocket and consequently the incomplete cytoplasmic rearranges and/or constricts and further flatten the columella[266,269−271].
-
During the past 30 years, seed development have been extensively investigated in Arabidopsis due to its importance in agriculture. In this review we briefly summarized major advances on some critical aspects during embryogenesis, endosperm development and seed coat formation. As for the signaling among these major structures during seed development we have reviewed recently[279]. The investigations mentioned in the present review greatly enhanced our understanding of the molecular mechanism underlying seed development, especially in some hot areas, such as zygotic genome activation, zygote asymmetric division, embryo pattern formation, parental contributions to the early embryogenesis, stem cell niche establishment at the embryo stage, the interaction between embryo proper and suspensor, programmed cell death of suspensor, endosperm development initiation and epigenetic regulation, the relationship between embryogenesis and endosperm development, and the seed coat development in relation to fertilization, embryo and endosperm development. The new findings updated our knowledge of seed development and importantly promote deeper investigations, as well as provide useful clues and tools for crop improvement.
However, some critical questions still await answers. For example, how the embryogenesis is triggered and what are the paternal and maternal roles in triggering embryogenesis. Although great efforts have been made in the past 30 years, the answer to the question still remains unclear. This answer is also related to another important question, how the parthenogenesis and other types of apomixis are triggered. Past research indicates that this is a field with great challenges, but recent findings in rice has thrown new light on the field and hopefully the mechanism underlying parthenogenesis will be clearly elucidated in the near future. Cell fate determination during embryogenesis is also a very complicated developmental issue, which may involve multilayer regulatory mechanisms, such as maternal influences, the cell type-specific gene expression, the interaction with neighboring cells, microenvironmental impacts. Embryo differentiation and organ formation are based on the cell fate determination process and the major players in the process may be distinct at different stages of embryogenesis for different tissue or organ formation, therefore, it is also a very challenging field. To overcome the difficulties the traditional strategy based on mutant analysis should be improved toward more positional accurate and more cell type-specific to get more detailed information. Cellularization is a critical step for endosperm development. however, the regulatory mechanism for the unique model of cytokinesis remains poorly understood. How the process is triggered, how the cell wall is generated at exact position and how the time course of the cellularization is related to seed size control are still puzzles waiting for further detailed elucidation. These examples illustrate that for understanding seed biology we still have long way to go in fundamental research.
In recent years, novel techniques have been quickly developed, which greatly accelerated the seed biological investigations. Single cell transcriptome has accumulated considerable amount of useful data of different cell types and tissues including zygote, embryos at different stages and endosperm. Careful analysis of these data may help us to find those key regulators in seed development. Improvement of the technology on single cell manipulations will enable us to select cells at specific positions in embryo, endosperm and seed coat for a well-targeted analysis. Living cell imaging together with molecular markers have turned out to be a very useful tool for monitoring the real dynamics of critical processes during seed development, especially, for following up the transient or compartmental behaviors of cells or proteins during cell-cell communications or interactions. In addition, seeds have been used as a platform for the so-called functional food production, which greatly enlarges the boarder of seed biology, and therefore, brings new tasks and challenges in this field. The synthesis and transportation of artificially-regulated storage materials in seeds during their maturation process, the regulatory mechanisms for the metabolism of these reserve substance, and the influence of over-loading of some beneficial substances in seeds on seed development requires more extensive investigations.
This work was supported by the Science and Technology Department of Hubei Province (2022CFA071) and the National Natural Science Foundation (Grant No. 32270360).
-
The authors declare that they have no conflict of interest. Mengxiang Sun is the Editorial Board member of Seed Biology. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
-
# These authors contributed equally: Xiaorong Huang, Peng Zhao, Xiongbo Peng
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Huang X, Zhao P, Peng X, Sun MX. 2023. Seed development in Arabidopsis: what we have learnt in the past 30 years. Seed Biology 2:6 doi: 10.48130/SeedBio-2023-0006
Seed development in Arabidopsis: what we have learnt in the past 30 years
- Received: 23 February 2023
- Accepted: 19 April 2023
- Published online: 05 June 2023
Abstract: Seed development is one of the extensively investigated fields of plant developmental biology due to its great agricultural and economic value. Arabidopsis thaliana as a model plant has been widely employed in seed development research over the past 30 years and these studies provide vast information involving various aspects of seed development, which have greatly enhanced our understanding of the critical processes of seed development. As it is very difficult to summarize all advances in this field, in the present review we have to focussed on key points concerning embryogenesis, endosperm development and seed coat formation. Molecular mechanisms regulating essential processes of seed development are briefly described to outline what we know to date, which have led and enhanced relevant investigations in the seed biology of crops.
-
Key words:
- Seed /
- Embryogenesis /
- Endosperm /
- Seed coat













