-
Dabing Zhang, a native of Huai'an City, Jiangsu Province (China), was born in 1967 into a rural family and, growing up, actively participated in farming activities. This background shaped his career path into agricultural research, specifically focusing on crop reproductive development and biosafety. Despite a relatively short 25-year career, Prof. Zhang made substantial and diverse contributions in various research areas, covering inflorescence development and spikelet specification; plant male reproduction and its adaptation to the environment; cytoskeleton dynamics and plant morphogenesis in response to environmental signals; and the molecular characterization and detection of genetically modified organisms (GMOs) in food and feed. In recognition of his exceptional work, he received the prestigious 2018 Prize in Agricultural Science from The World Academy of Sciences (TWAS) for his significant contribution towards understanding the molecular mechanisms involved in morphogenesis of inflorescence, flowers, and anthers in higher plants, with a particular focus on rice.
This commemorative article comprehensively summarizes and organizes Zhang's scientific achievements, which serve as a foundation for future advances in these fields.
Inflorescence, spikelet, and flower development and specification -
Reproduction is a crucial step in the plant life cycle that directly affects yield in agricultural crops[1−3]. The molecular regulatory mechanisms governing floral, spikelet, and inflorescence development have therefore garnered significant attention from botanists and crop scientists (Fig. 1a). The classical ABCDE model of flower organ development — the 'quartet model' — was formulated based on studies conducted in dicots Arabidopsis, snapdragons, and petunias. According to this model, establishment and development of the outermost whorl of floral organs (sepals) is regulated by class A genes; the second whorl (petals) by both class A and B genes; the third whorl (stamens) by class B and C genes; and the fourth whorl (carpels) by class C genes. Additionally, class C and D genes redundantly contribute to the establishment and development of ovules within carpels, while class E genes contribute to flower identity determination and participate in the establishment and development of each whorl[4,5].
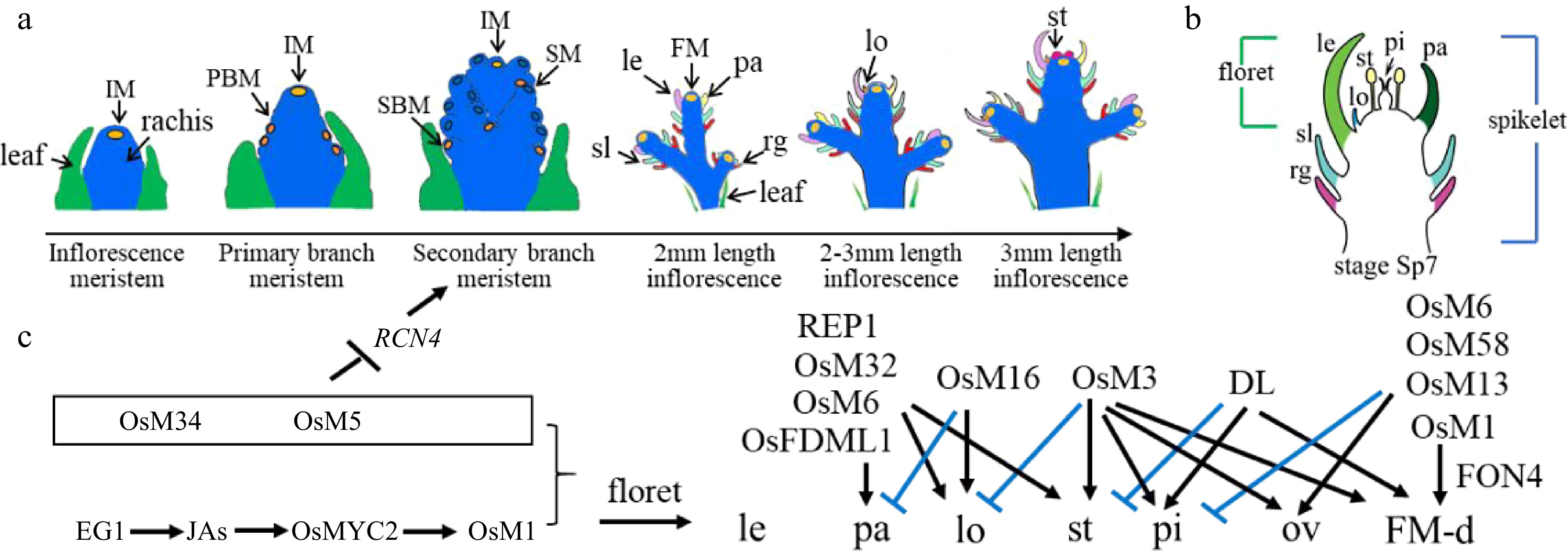
Figure 1.
Rice inflorescence and spikelet development. (a) Schematic diagram of rice inflorescence development from the inflorescence meristem to stamen stages, showing leaf cells (green), rachis cells (blue), meristem cells (yellow), and other colors as annotated: FM, floret meristem; IM, inflorescence meristem; le, lemma; lo, lodicule; pa, palea; PBM, primary branch meristem; rg, rudimentary glume; SBM, secondary branch meristem; sl, sterile lemma; SM, spikelet meristem; st, stamen. (b) Schematic diagram of a rice spikelet, comprising two sterile lemmas and rudimentary glumes, and one floret. pi: pistil. (c) Genes that were cloned and studied by Prof. Zhang's group. Black color represents positive regulation or activation, while blue color represents negative regulation or repression. JA, jasmonic acid; OsM, OsMADS; ov: ovule; FM-d: flower meristem determinacy.
Subsequent biochemical and molecular biological investigations have elucidated the molecular foundation of the ABCDE model, which proposes that MADS-box proteins encoded by the class A–E genes form specific tetramers to regulate the expression of target genes, thereby exerting control over the establishment and development of each whorl. Notably, class E proteins act as the 'glue' and can form heterotetramers with class A, B, C, and D proteins[6].
Unlike the floral structure of typical dicot flowers, grass flowers contain a palea and lemma in the outer whorl, whose characteristics and origin remain the subject of ongoing debate, and lead to some questions over the relationship between monocot and dicot flower organs.
To address these questions, Prof. Zhang explored the genetic and biochemical bases of inflorescence and spikelet development in rice, a model grass species. Wild-type rice spikelets contain one terminal floret accompanied by two whorls of subsidiary organs comprising rudimentary glumes and sterile lemmas (Fig. 1b). Rice florets consist of one lemma, one palea, two lodicules next to the lemma, six stamens, and one pistil.
With his group, Prof. Zhang constructed a rice mutant library using ethyl methanesulfonate (EMS) mutagenesis and 60Co γ-ray irradiation[7]. This library has served as a rich analytical resource, and his staff and students have used it to clone and characterise the functions of B, C, D, and E class orthologous genes, as well as to elucidate the role of other transcription factors (TFs) in rice floret development (Fig. 1c).
Floral organ number 4
-
The floral organ number 4 (fon4) mutants exhibit an increased number of floral organs and primary rachis branches. This phenotype arises due to enlarged shoot apical meristems (SAMs) caused by mutation of the small secreted FON4 protein, the ortholog of the Arabidopsis CLAVATA3 (CLV3); both orthologs function by inhibiting meristem size during vegetative and reproductive development, implying a conserved CLV-like pathway in both rice and Arabidopsis[8]. While FON4 transcripts predominantly accumulate in a specific group of cells located at the apex of the SAMs, treatment with FON4 peptide does not impact root apical meristem (RAM) growth. This does differ from CLV3 effects in Arabidopsis, suggesting at least some functional divergence in the CLV3/ENDOSPERM SURROUNDING REGION (ESR)-related (CLE) motifs of Arabidopsis CLV3 and rice FON4.
In cooperation with Prof. Jian Xu (Department of Plant Systems Physiology, Radboud Institute for Biological and Environmental Sciences, Radboud University, The Netherlands), Prof. Zhang's group discovered that the rice FON2-LIKE CLE PROTEIN2 (FCP2p) terminates RAM activity and impairs late metaxylem formation by suppressing expression of the rice QUIESCENT-CENTER-SPECIFIC HOMEOBOX (QHB) gene. QHB is considered to be a counterpart of the Arabidopsis WUSCHEL (WUS)-RELATED HOMEOBOX 5 (WOX5) gene, and its expression is affected in both quiescent center and late metaxylem cells. Remarkably, this regulatory network is conserved in Arabidopsis, providing a new perspective on the role of CLE–WOX signaling in RAM maintenance and vascular tissue development[9].
Furthermore, a recent study found that FON4 with the C, D, and E floral homeotic genes collaboratively contribute to the specification of FM activity in floral development[10]. Moreover, research conducted by Prof. Qian Qian's lab (State Key Laboratory of Rice Biology, China National Rice Research Institute, China) revealed that FON4 may synergistically interact with various partners, including the HD-ZIP III TF LATERAL FLORET 1 (LF1); YABBY TF TONGARI-BOUSHI1 (TOB1); AP2 TFs MALE AND FEMALE STERILITY1 (MFS1) and SUPERNUMERARY BRACT (SNB); and MADS TF OsMADS22. These interactions contribute to the regulation of spikelet meristem determinacy and retention of single floret spikelets in rice[11]. As such, these findings present potential for the development of rice cultivars with multi-floret spikelets.
Genes encoding MADS-box homeodomain TFs
-
A complex regulatory network in determining rice floral meristems was revealed on discovery of an interaction between TFs encoded by rice floral homeotic genes, OsMADS3 (C class), OsMADS13 (D class), and DROOPING LEAF (DL, a YABBY TF), to determine ovules and floral meristem identity. OsMADS3 and DL also work together to terminate the floral meristem, while DL and OsMADS13 may function in the same pathway to determine the identity of carpel/ovules and floral meristems[12].
The AGAMOUS-LIKE6 (AGL6) MADS-box genes (OsMADS6 and OsMADS17), along with the E class genes that include the LOFSEP (OsMADS1, OsMADS5, and OsMADS34) and SEP3 (OsMADS7 and OsMADS8) clades, were also identified as key regulators of rice meristem activity. OsMADS6 positively regulates the expression of OsMADS7, OsMADS8, OsMADS3, OsMADS16/SUPERWOMAN1 (SPW1), and OsMADS58. Furthermore, OsMADS6, OsMADS1, OsMADS13, and DL redundantly regulate floral meristem activity and identity, and combinational double mutants develop severely indeterminate floral meristems[13,14]. However, it remains to be elucidated why there appear to be so many redundant E class genes and whether any of them have specific functions in meristem activity.
Roles of E class MADS-box TFs in flower and inflorescence development
-
Recent research has revealed that HvMADS1 in barley plays a crucial role in inhibiting inflorescence branching to maintain inflorescence (spike) morphology under high temperature conditions. While unbranched at low temperatures, the Hvmads1 spike becomes branched at high temperatures. Further investigations demonstrated that HvMADS1 promotes the expression of cytokinin oxidase/dehydrogenase 3 (CKX3), increased at higher temperatures, whose encoded protein degrades the cytokinin phytohormone. In the Hvmads1 mutant with reduced CKX3 expression, heat therefore results in increased cytokinin levels that promote branching, corroborated by knockout of the CKX3 gene that causes a similar branched spike phenotype under high temperature conditions[15]. This finding presents compelling evidence linking variations in inflorescence branching among grass crops to temperature conditions. However, the conservation and significance of the MADS1 mechanism in regulating cytokinin content across rice and other grass species remain uncertain.
Research conducted by Prof. Zhang's team further confirmed the functional differentiation of rice E class LOFSEP genes OsMADS1, OsMADS5, and OsMADS34. Investigation into the functions and genetic interactions of OsMADS1 and OsMADS34 revealed that these genes co-regulate the development of rice floral organs — lemma, palea, stamens, and pistils[16,17]. By contrast, allelic mutants of OsMADS5 exhibited a weak phenotype, implying a potential minor role of this gene in spikelet development. The team constructed Osmads1/5/34 (lofsep) triple mutants and all possible double mutant combinations, observing a gradual and dosage-dependent transformation of spikelets into leaf-like structures in lofsep single, double, and triple mutants[18]. These genetic and molecular insights into the overlapping regulation of rice spikelet traits and floral meristem determination revealed that, unlike in dicot plants, LOFSEP genes in grasses regulate the development of the inner three whorls of spikelet organs by transcriptionally activating B, C, AGL6, and SEP3 class genes. Moreover, the study showed that LOFSEP and SEP proteins share conserved characteristics, enabling interaction with other MADS-box proteins to form complexes[18].
A further recent study revealed the collaborative regulation of OsMADS5 and OsMADS34 during rice inflorescence development. OsMADS34 accumulates in the early stages of inflorescence development, while OsMADS5 expression initiates during stage four (In4) development. OsMADS5 exhibits higher expression in the secondary branch and floral meristems, with clear overlap with OsMADS34 expression domains. Molecular experiments confirmed the direct binding of OsMADS5 and OsMADS34 to the RCN4 promoter to co-suppress RCN4 expression. This process limits spikelet branching while promoting the transformation of floral meristem characteristics[19].
Interaction with jasmonic acid phytohormone regulation
-
The identification and characterization of two mutants exhibiting additional glumes revealed a novel signaling pathway involved in rice floret initiation and meristem identity determination[20]. Abnormal spikelets in extra glume1-3 (eg1-3) single mutants and eg2-1D exhibited changes in floral organ identity, number, and defective floral meristem determinacy. Moreover, the double eg1-3 eg2-1D mutants grew additional ectopic floral primordia, glume-like structures, or double floral meristems within a single spikelet, indicating abnormalities in floral meristem indeterminacy.
Molecular analyses revealed that EG1 encodes a lipase involved in jasmonic acid (JA) biosynthesis, while EG2 (also known as OsJAZ1) acts as a repressor of the JA response and interacts with JA receptor OsCOI1b. Prof. Zhang's research thus demonstrated that JAs, their content, and signaling transduction play a crucial role in floret initiation by activating the expression of OsMADS1, OsMADS7, and OsMADS8. This pathway has been further confirmed by other research groups, which also found evidence supporting the involvement of genes involved in JA biosynthesis (OsPEX5 and OsOPR7)[21], JA receptors (OsCOI1a and OsCOI1b)[22], and a JA-responsive gene (OsMYC2)[21]. Although genetic analysis has shown that OsMADS1 and OsMADS3 are crucial for maintaining floral meristem activity and specifying organ identity, and OsMADS1 and OsMADS58 are important in regulating floral meristem determinacy and suppressing spikelet meristem reversion[23], the exact mechanism by which rice OsMADS1 regulates floret meristem identity remains elusive.
Resolving whorl identity in monocots and dicots
-
Since the function of B and C class genes are highly conserved in Arabidopsis and rice, Prof. Zhang's research hypothesized that the palea is the counterpart of the sepal in eudicots, and the lemma is homologous to the bract[24]. This opinion is consistent with the study on the LONG STERILE LEMMA (G1)[25], also known as the ELONGATED EMPTY GLUME (ELE)[26], and is further supported by the function of LF1 in specifying sterile lemma identity[27]. In contrast to the single mutant of B class (OsMADS16/SPW1) or C class (OsMADS3, OsMADS58) genes, the double mutant combinations, spw1-1 osmads3-4 and spw1-1 osmads58, grew an additional whorl consisting of six glume-like structures exhibiting palea identity at the location of the wild-type stamens, confirming that B and C class genes in rice play a pivotal role in suppressing indeterminate growth within the floral meristem, especially in whorl 3 primordia.
A recent single-cell RNA sequencing analysis investigated the developmental trajectories of rice floret and inflorescence meristems. The findings revealed a significant disparity between the cellular transcriptomic atlas of the palea and the lemma. Furthermore, it indicated that a complete rice flower is composed of the palea, lodicule, stamen, and pistil[28]. In conclusion, the genetic and molecular evidence presented above unequivocally establishes the homology between the sterile lemma and the lemma, confirming their identity as a bract. Additionally, the palea is identified as a sepal-like structure within the rice floret. The mechanisms by which the palea establishes its distinct identity and undergoes morphogenesis are worthy of further investigation.
Existing evidence suggests that MADS TFs OsMADS15, OsMADS32, OsMADS6, and the TCP TF REP1 might play significant roles in regulating rice palea morphogenesis. Mutants, such as degenerative palea (dep)/Osmads15, retarded palea1 (rep1), and depressed palea1 (dp1), show delayed or even arrested development of the body of palea (BOP), while the marginal region of palea (MRP) remains unaffected[29−31]. The chimeric floral organs1 (cfo1)/Osmads32 mutant displays a transformation of the MRP into the BOP[32,33]. In the mosaic floral organs1 (mfo1)/Osmads6 mutant, the MRP undergoes transformation into the BOP and expands outward, resembling the lemma with five vascular bundles. OsMADS6 directly regulates the expression of the FACTOR OF DNA METHYLATION LIKE 1 (OsFDML1), and the Osfdml1 mutant palea resembles that of the mfo1/Osmads6 mutant. Transgenic plants overexpressing OsFDML1 or OsMADS6 also display abnormal palea development, characterized by a loss of delineation between BOP and MRP cells[34]. This chimeric structure leads to the outward growth of the BOP, indicating that the MRP normally restricts BOP growth. These observations suggest that the development of rice palea, especially the MRP, is finely regulated; the specific molecular mechanisms warrant further investigation.
Plant male reproduction and its adaptation to the environment -
Plant male reproduction occurring in the stamen is essential for successful seed setting in flowering plants. The stamen consists of the anther and the filament; the former produces pollen while the latter connects the anther to the central floral axis and provides a conduit for water and nutrients. Stamen development initiates with the floral primordia to form the filament and the anther. Meristematic cells in the anther divide and differentiate into pollen mother cells, which finally generate the male gametophyte (pollen) and the multiple somatic cell layers that form the surrounding maternal anther walls (epidermis, endothecium, middle layer, and tapetum). The development of viable pollen and its subsequent release from the anther is a complex process that requires tight regulatory control, and is remarkably conserved in model plants, such as Arabidopsis and rice[35−37]. The successful formation and ultimate release of the pollen requires timely formation and degeneration of anther somatic cell layers and smooth communication between anther cell layers and the developing pollen[36]. One critical somatic cell layer is the tapetum, whose degradation through programmed cell death (PCD) during meiosis provides not only constructive and nutritive components for pollen wall formation and accumulation of intracellular starch and lipids, but also signal molecules for pollen development[35,38−40]. During tapetum PCD, microspores form primexine, the template for sporopollenin precursor deposition and assembly, which forms an elaborate, three-layered wall — outer exine, inner intine, and pollen coat, also known as tryphine[38−40] — that protects the mature pollen grains.
Prof. Zhang contributed significantly to the molecular understanding of male reproduction in both rice and Arabidopsis using integrated genetic, biochemical, molecular, anatomical, and cytological approaches (Fig. 2).
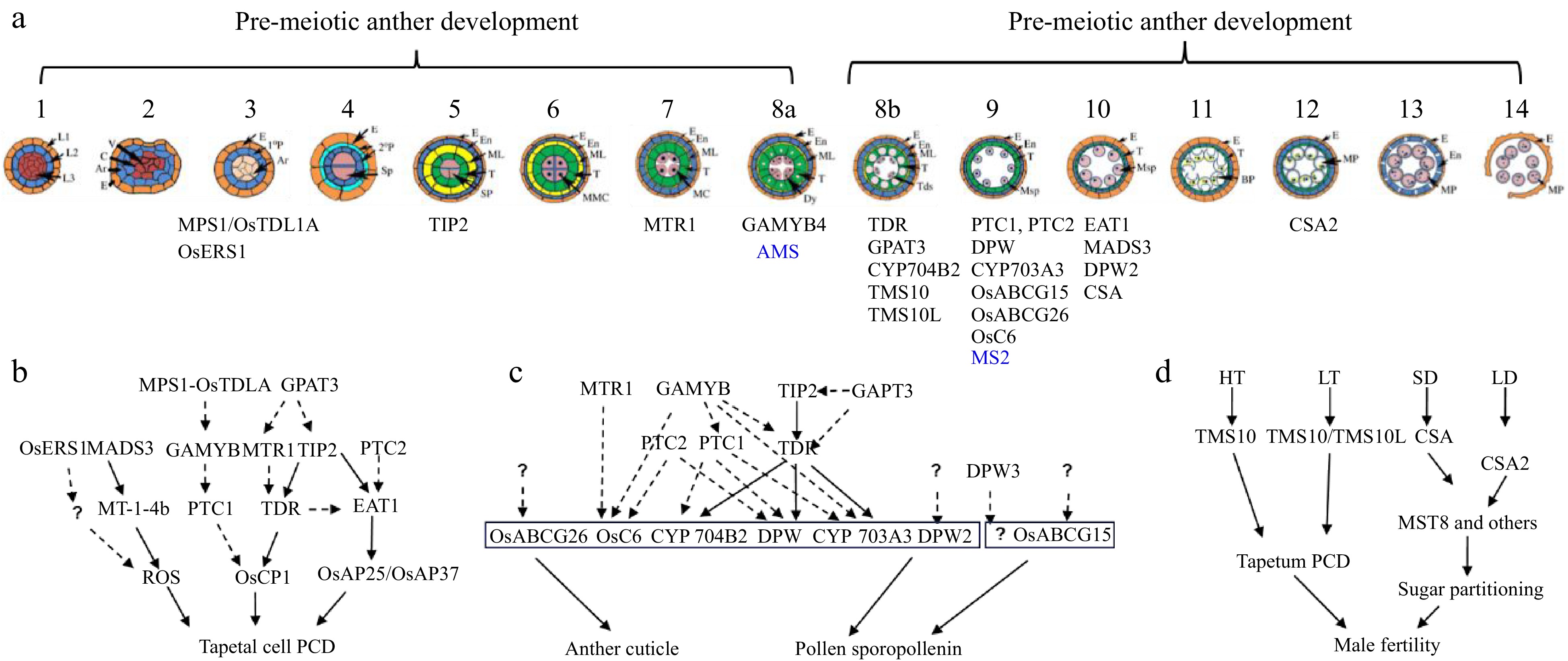
Figure 2.
Anther and pollen development and its adaptation to the environment. (a) Schematic overview of anther development in dicot and monocot plants. Genes identified by Prof. Zhang with a role in anther development are shown (blue for Arabidopsis and black for rice). (b) Regulatory aspects of anther development. (c) Pathways and networks related to rice tapetal programmed cell death (PCD), and anther cuticle and pollen sporopollenin development, all identified by Prof. Zhang. Solid arrows represent information obtained from biochemical binding assays, while dashed arrows indicate data from transcriptional analysis. Question marks indicate ongoing gaps in knowledge. (d) Prof. Zhang has identified two molecular pairs, TMS10/TMS10L and CSA/CSA2, that mediate rice responses to high/low temperature (HT, LT) and long-short day length (LD, SD), respectively.
Premeiotic anther development
-
Intracellular communication and differentiation of meristematic cells are essential processes for successful premeiotic anther development and subsequent pollen formation, yet the underlying molecular mechanisms remain largely unknown. In this area, in 2012, Prof. Zhang's group revealed the crucial role of a plasma membrane–localized rice fasciclin glycoprotein, MICROSPORE AND TAPETUM REGULATOR1 (MTR1), in communication between sporophytic and reproductive cells. Male reproductive cells secrete MTR1, which acts as an extracellular adhesion molecule to regulate cell-to-cell communication between reproductive and adjacent somatic cells[41].
In 2016, his research revealed the role of a cell surface-localized leucine-rich repeat receptor-like kinase (LRR-RLK), MSP1 (MULTIPLE SPOROCYTE1), in determining early anther cell fate. MSP1 interacts with its ligand, OsTDL1A (TPD1-like 1A), to specify the cell identity of anther wall layers and microsporocytes; single and double mutants lack middle layers and tapetal cells and have more microsporocytes. MSP1 and OsTDL1A exert their effects by modulating redox status via the TGACGTCA cis-element-binding protein OsTGA10 and plant-specific CC-type glutaredoxin OsGrx_I1[42].
In 2018, Prof. Zhang's group also elucidated the critical role of the cytosol- and mitochondria-localized glutamyl-tRNA synthetase (GluRS), OsERS1, in anther cell division and patterning. OsERS1 fulfills its regulatory function by affecting protein synthesis, amino acid–derived metabolism, and cellular redox hemostasis[43]. Another noteworthy mention is TDR INTERACTING PROTEIN2 (TIP2), a basis helix-loop-helix (bHLH) protein, which was found to directly regulate the expression of genes encoding two other bHLH proteins, Tapetum Degeneration Retardation (TDR) and ETERNAL TAPETUM 1 (EAT1). TIP2 also interacts with TDR, affecting late anther development, suggesting that these three bHLH proteins play a central role in regulating distinct steps of plant cell-type determination[44]. These results unveiled one of the mysteries in biology regarding mechanisms controlling the specification and communication of the tapetum and microsporocyte cells[45], and laid a solid foundation for plant breeding via manipulation of premeiotic anther development.
Post-meiotic anther development
Tapetum PCD
-
In flowering plants, PCD in the tapetum triggers its degeneration during the late stages of pollen development to provide cellular contents and signal molecules supporting pollen wall formation and pollen maturation. Findings from Prof. Zhang's group significantly enriched our understanding of the molecular bases regulating tapetum PCD in plants. His group isolated and characterized several TFs that are important regulators of tapetum PCD, including:
• TDR, which promotes tapetal cell PCD by direct targeting OsCP1 (a cysteine protease) and OsC6 (a protease inhibitor)[46]; it is also important for several basic biological processes, such as aliphatic lipid metabolism and rice pollen development[47];
• EAT1, interacting with and acting downstream of TDR to promote tapetum PCD by direct regulating aspartic proteases OsAP25 and OsAP37[48];
• The homeotic C class protein, MADS3, which modulates tapetum PCD, at least in part, by regulating anther reactive oxygen species (ROS) balance through the ROS-removal protein MT-1-4b and relevant metabolic pathways[49,50];
• A plant homeodomain (PHD)-finger protein PERSISTENT TAPETAL CELL1 (PTC1) that promotes tapetum necrosis-like cell death in a unknown manner[51]; and
• A nuclear localized AT-hook protein PTC2 that has similar function, by an equally unknown mechanism, to PTC1 in tapetum PCD[52].
Notably, although detailed molecular bases underlying their function in tapetum PCD have not been provided for all abovementioned TFs, these studies highlight their important roles in anther and pollen development, and provide novel insights into plant male reproduction (Fig. 2b).
Anther cuticle and pollen sporopollenin formation
-
One of the greatest contributions of Prof. Zhang's group to plant reproductive biology is the systematic studies on genes and networks that regulate the biosynthesis, modification, transportation, and assembly of anther cuticle and pollen sporopollenin, two important protective envelopes for pollen. He and his group identified and characterized many genes encoding the involved enzymes, transporters, and TFs (Fig. 2c).
Biosynthesis of lipidic precursors
-
Prof. Zhang's work on Defective Pollen Wall (DPW) in rice revealed, for the first time, that fatty alcohols play key roles in anther cuticle and pollen sporopollenin biosynthesis. The plastid-localized DPW converts fatty acids to fatty alcohols to facilitate their export from the plastid to the cytosol for incorporation into the cuticle and sporopollenin[53]. His work also confirmed that this DPW-mediated plastidial pathway for fatty alcohol production is also conserved in Arabidopsis[54], highlighting the essential role of fatty alcohols in male reproductive development. Additional work revealed that DPW3, encoding an alpha integrin-like protein, is also essential for anther cuticle and pollen sporopollenin development via its effects on primexine matrix formation, plasma membrane undulation, callose wall deposition, and Ubisch body formation[55].
Identification of a rice endoplasmic reticulum–localized enzyme, glycerol-3-phosphate acyltransferase 3 (OsGPAT3), from the 60Co γ-ray irradiation-generated mutant population in rice[7], also provided new insights into the function of glycerolipid biosynthetic enzymes in male reproduction. OsGPAT3 is monocot-specific, playing different roles in male reproduction from dicot counterparts; and osgapt3 mutant shows defective anther cuticle, abnormal tapetum development and degeneration, degenerated pollen with defective exine, and complete male sterile[56].
Modifications of lipidic precursors
-
The isolation and characterization of two cytochrome P450 family member proteins in rice underlay recognition of the essential role of lipid modification in anther cuticle and pollen sporopollenin biosynthesis. CYP704B2 catalyzes the production of ω-hydroxylated fatty acids, with a conserved role in plant male reproductive development from mosses to angiosperms[57]. CYP703A3 is an in-chain hydroxylase that catalyzes only lauric acid, preferably generating 7-hydroxylated lauric acid, demonstrating the importance of this pathway for male reproductive development; interestingly, TDR directly regulates CYP703A3 expression[58]. Both mutants show defective anther cuticle, pollen sporopollenin biosynthesis and male sterility[57, 58].
Transport of lipidic precursors
-
Prof. Zhang's work also identified two types of transporters in rice, explaining how various metabolites produced in tapetum are allocated between different anther tissues. One such a transporter is plant lipid transfer protein (LTP) OsC6. This monocot-specific protein has lipid-binding activity; is indispensable for the development of lipidic orbicules (Ubisch bodies) and pollen sporopollenin; is involved in anther cuticle and sporopollenin biosynthesis; and is regulated directly by TDR[59]. The other type of transporters are ATP binding cassette G family members OsABCG26 (Arabidopsis ortholog ABCG15) and OsABCG15 (Arabidopsis ortholog ABCG11). OsABCG26 and OsABCG15 collaboratively regulate rice male reproduction by affecting the transport of lipid precursors from tapetal cells: OsABCG26 transports lipidic molecules for both anther cuticle and sporopollenin formation; while OsABCG15 transports molecules mainly for sporopollenin development[60, 61].
Assembly of lipidic precursors
-
Functional characterization of DPW2, a cytoplasmic hydroxycinnamoyl-CoA:ω-hydroxy fatty acid transferase, showed that aromatic acids are essential for the assembly of lipidic precursors to form anther cuticle and pollen sporopollenin, demonstrating that DPW2 plays a fundamental role in plant reproductive development by polymerizing lipidic precursors to form anther cuticles and pollen sporopollenin[62]. This work also highlighted equal roles of aromatic lipids to aliphatic lipids in male reproduction.
Anther sugar partitioning
-
Functional characterization of Carbon Starved Anther (CSA) in rice pinpointed the importance of sugar partitioning between sink and source tissues for anther development and male reproduction. CSA is an R2R3 MYB TF that direct regulates the expression of MST8, encoding a monosaccharide transporter, showing that CSA is a key transcriptional regulator for sugar partitioning in rice male reproductive development[63]. Notably, brassinosteroid (BR) phytohormones promote pollen and seed development in rice by direct regulating CSA expression, connecting BRs and sugar partitioning in plant reproduction[64].
Regulation of anther and pollen development
Transcriptional regulation
-
Work by Prof. Zhang and his colleagues identified many important TFs involved in regulation of plant male reproductive development (Fig. 2b, c), including:
• TIP2, alone involved in pre-meiotic anther development, and, together with TDR in late anther development[44];
• TDR[46,47], EAT1[48], MADS3[49], PTC1[51], and PTC2[52] involved in tapetum PCD;
• CSA, involved in sugar partitioning[63];
• GAMYB, a gibberellic acid (GA)–inducible TF that, together with UNDEVELOPED TAPETUM 1 (UDT1), involved in the regulation of rice early anther development[65]. The msp1-4 mutation results in reduced expression of UDT1 and GAMYB, implying that MSP1 likely acts upstream of UDT1 and GAMYB in male reproductive development[66];
• ABORTED MICROSPORES (AMS), the ortholog of TDR in Arabidopsis. A bHLH TF, involved in tapetum and pollen development. AMS can directly regulate genes involved in anther cuticle and pollen sporopollenin formation; and interacts with other two bHLH proteins (AtbHLH089 and AtbHLH091) and the ATA20 protein, forming a complex to co-regulate anther and pollen development[67,68].
From basic research to advances in breeding -
Zhang's work enriched not only our theoretical understanding of plant male reproductive biology, but has also contributed significantly to improvements in hybrid rice breeding, the most effective strategy to significantly increase rice yield.
Adaptation to temperature
-
In rice, Zhang and his group identified a molecular switch controlling the adaption of plant male reproduction to fluctuating temperatures, which revealed new molecular mechanisms by which plants control reproductive development in response to environment. This molecular switch consists of two LRR-RLKs, Thermo-Sensitive Genic Male Sterile 10 (TMS10) and its close homolog TMS10-Like (TMS10L), which redundantly maintain anther and pollen development at normal temperatures (Fig. 2d). TMS10 alone mediates male reproduction at high temperatures, while TMS10 and TMS10L together mediate male reproduction at low temperatures[69]. This work provided the rice community with a useful genetic resource for the development of new hybrid seed production systems.
Adaptation to photoperiod
-
The functional characterization of CSA2, another MYB TF, by Prof. Zhang's group revealed another molecular switch modulating male reproduction in response to photoperiod (Fig. 2d). This study revealed that rice evolved at least two MYB proteins, CSA and CSA2, to regulate sugar partitioning from source leaf tissues to the sink anther tissues under short-day and long-day conditions, respectively[70]. After comparing transcriptomic profiles of csa mutant to wild-type (WT) leaves and anthers under different photoperiods, Prof. Zhang's research identified eight direct targets of CSA involved in sugar metabolism and transport[71].
To further identify key genes and regulatory networks affecting pollen maturation in rice anthers in response to different day length, Prof. Zhang and his colleagues performed transcriptomic studies on Stage 11 anthers collected at different times during the day to create a new resource for identifying environmentally sensitive genes regulating male reproductive development for use in rice improvement[72].
These studies provide not only insights into molecular mechanisms controlling male fertility in response to changing photoperiod but also genetic resources for resilient rice breeding. To overcome intrinsic problems in current male sterile line-dependent hybrid rice technologies, Prof. Zhang and his colleagues introduced the csa mutation into indica and japonica rice, creating lines in both varieties that were male-sterile under short-day conditions but male-fertile under long-day conditions. Most importantly, F1 plants of csa and a restorer line JP69 show hybrid vigor, allowing establishment of a stable two-line hybrid system[73], an advance with high potential for significant impact on agriculture.
New insights into the role of the cytoskeleton in rice development and environmental responses -
The architecture of flowering plants exhibits remarkable diversity due to variations in organ morphology. The shape of individual plant organs results from the shape and structure of cells within them, with specialized morphology maintained by the microtubule and microfilament cytoskeletal systems. The plant cytoskeleton, primarily composed of microtubules and actin filaments (AFs), is a highly dynamic structure in plant cells. Extensive research has highlighted its crucial role in various intracellular processes, such as cell division, movement, morphogenesis, and signal transduction.
Forward genetic screening by Prof. Zhang and his research team was used to identify a mutant displaying abnormal rice architecture, characterized by reduced plant height, root skewing, smaller seed size, and other traits (Fig. 3). The causative gene, Rice Morphology Determinant (RMD, Os07g0596300), encoding a type II formin, is the first formin protein verified to play significant roles in rice development. As an actin-binding protein, it regulates AF and tubulin dynamics, acting as a scaffold between cellular organelles and the cytoskeleton[74,75]. The resulting new insights into RMD's involvement in shoot and root system architecture determination, pollen tube elongation, light-mediated shoot gravitropic response, and secondary cell wall formation in the stem have significantly advanced our understanding of the cytoskeleton's contribution to rice development and response to environmental cues. Ongoing efforts by the team are generating new knowledge in this field, with further breakthroughs anticipated.

Figure 3.
rmd mutants exhibit pleiotropic defects compared with wild type (WT) plants[75]. (a) Seedlings; d, days after germination. Bar = 8 mm. (b) Rice plants at heading stage. Bar = 20 cm. (c) Panicle and rachis of plants at heading stage. Bar = 2 cm. (d) Individual flowers from plants at heading stage. Bar = 2 mm. (e) Seeds. Bar = 2 mm.
Role of RMD in pollen tube growth
-
The pollen tube serves as a conduit for transporting sperm cells to the ovules, facilitating double fertilization in angiosperms. Prof. Zhang and colleagues elucidated the underlying mechanism through which RMD promotes the arrangement of actin filaments in pollen tube growth. Functional loss of RMD impeded the rate of pollen tube initiation (germination) and growth, both in vitro and in vivo. The localization of RMD at the apex of the rice pollen tube plays a pivotal role in promoting pollen tube growth by establishing polarity and organizing F-actin. The rmd mutants exhibited an irregular F-actin pattern with disrupted apical actin density and shank longitudinal cable arrangement. The orientation and properties of the F-actin array, guided by RMD, critically influence the deposition of cell wall components, e.g., pectin, and the pattern and speed of cytoplasmic streaming[76]. This study demonstrated the essential role of RMD in spatial regulation of rice pollen tube growth by modulating the organization and orientation of the F-actin array (Fig. 4).
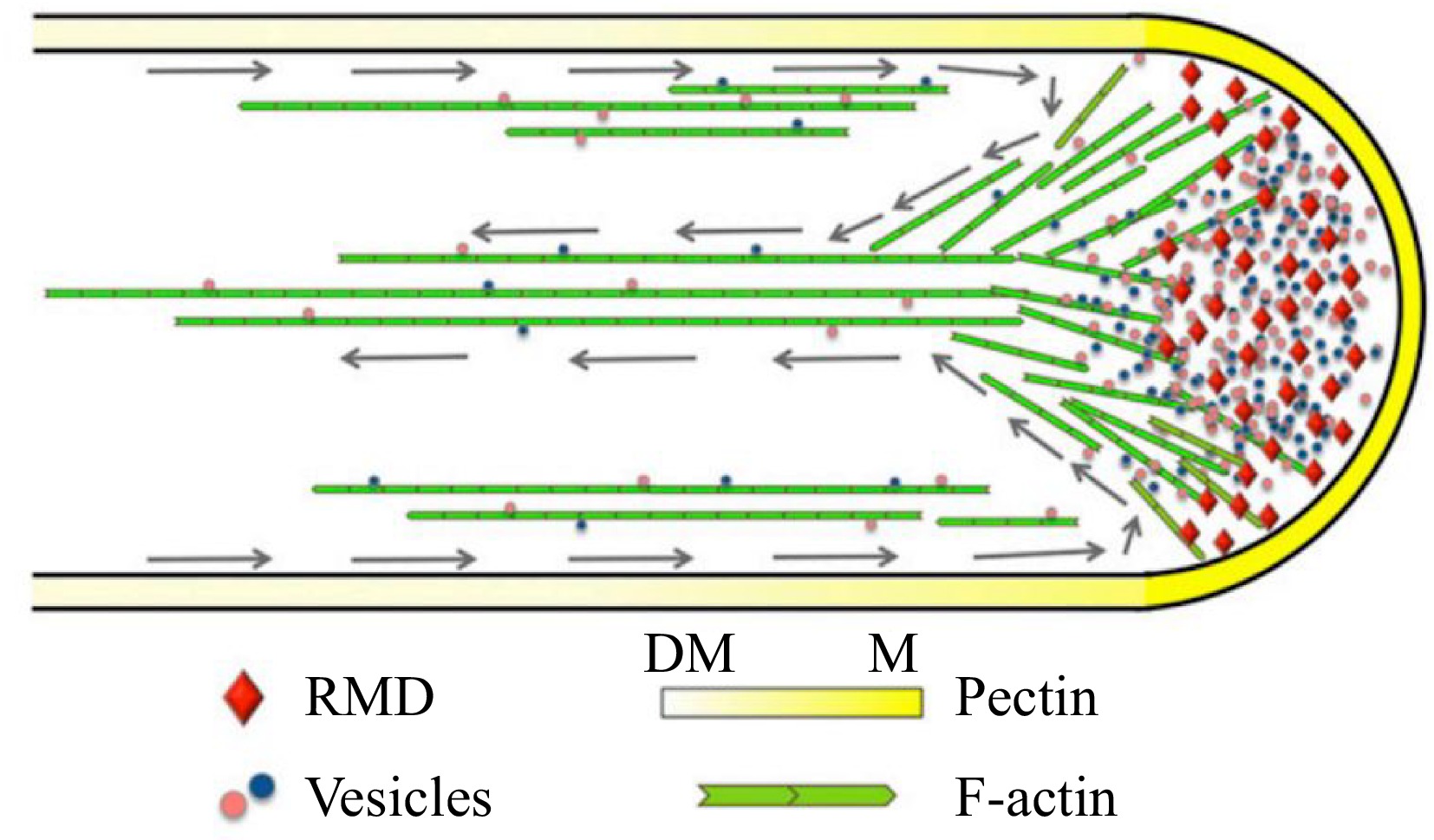
Figure 4.
Model depicting the function of RMD in rice pollen tube growth[76]. RMD serves as a crucial regulatory factor that plays a significant role in the maintenance of the spatial arrangement of cytoplasmic streaming, which is vital for the determination of pectin distribution and subsequent tip growth of the rice pollen tube. Gray arrows trace the reverse-fountain cytoplasmic streaming pattern. DM, demethyl-esterified; M, methyl-esterified.
Role of RMD in determining root angle in response to soil nutrients
-
Root angle plays a significant role in nutrient uptake, particularly for phosphate, which tends to accumulate in the top layer of soil. In response to decreased phosphate levels, roots tend to grow at a shallow angle, which compromises low phosphate tolerance and other elements of plant fitness. Consequently, the identification of genes and mechanisms that regulate root angle is of utmost importance in plant breeding. With Prof. Malcolm Bennet (University of Nottingham, UK), Prof. Zhang revealed that RMD is upregulated in response to low external phosphate, and that RMD localizes and links AFs to gravity-sensing organelles called statoliths. The rmd mutants exhibited steeper crown root growth angles, unresponsive to phosphate levels, and demonstrated a faster gravitropic response due to more rapid statolith movement (Fig. 5)[77]. This study provides novel mechanistic insights into the regulatory mechanisms used by plants to modulate crucial elements of the gravitropic machinery, enabling them to adjust the architecture of their root systems according to the availability of soil nutrients. Furthermore, it identifies further potential molecular targets for plant breeding purposes.
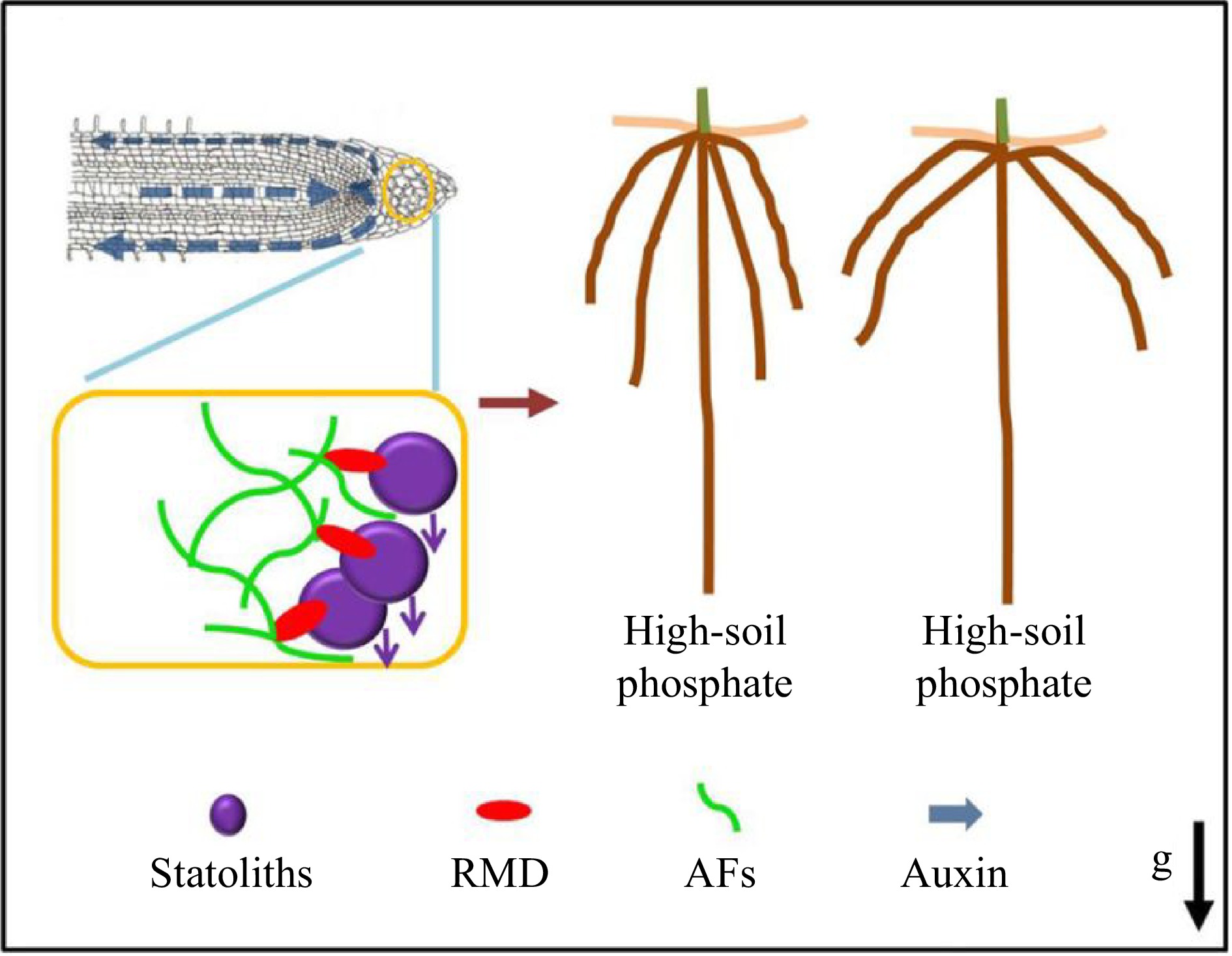
Figure 5.
RMD-dependent regulation of root crown angle by phosphate[77]. Prof. Zhang and collaborators proposed a model whereby low soil phosphate conditions and higher RMD levels in columella cells promote stronger interactions between AFs and statoliths that delay statolith sedimentation, resulting in less robust auxin-driven root gravitropic response and shallower crown root angle. In contrast, lower RMD levels in response to high soil phosphate ultimately result in a steeper crown root angle.
The role of RMD in light signaling and gravity perception
-
Light and gravity play crucial roles in the orientation of plant stems for optimal growth and development, the physical movement of which is tightly bound to the organization and dynamics of the actin cytoskeleton and actin-binding proteins. Nevertheless, the involvement of the actin cytoskeleton in shoot negative gravitropism is a subject of ongoing debate.
Prof. Zhang and his research team have documented that RMD facilitates reorganization of the actin cytoskeleton in rice shoots, closely linked to the rice shoots' capacity to respond to negative gravitropism. Specifically, rmd mutant shoots grown under light conditions exhibited agravitropic phenotypes, while etiolated rmd shoots demonstrated typical negative shoot gravitropism. Moreover, this study demonstrated that RMD plays a crucial role in maintaining an actin configuration that facilitates the movement of statoliths in endodermal cells responsible for gravity sensing. Additionally, RMD is involved in ensuring proper distribution of auxin in shoots grown under light conditions, but not in those grown in darkness. The expression of RMD is under diurnal control and directly suppressed by the phytochrome-interacting factor-like protein OsPIL16. Consequently, overexpression of OsPIL16 results in defects in gravity-sensing and actin patterning, which closely resemble the phenotypic characteristics of the rmd mutant[78]. These findings provide valuable insights into the intricate mechanism that connects light signaling and gravity perception, ultimately contributing to the straight upward growth of rice shoots.
Molecular characterization and detection of genetically modified organisms -
Since 1996, China has made significant advancements in the development and approval of industrial production and application of a wide range of genetically modified (GM) crops. Nonetheless, the safety of GM crops and their products has become a major concern for the general public. To address this issue, Prof. Zhang and his group have undertaken thorough research on the key challenges associated with the risk assessment and management of GM organisms (GMOs). This research has focused on crucial areas including the identification and analysis of molecular characteristics, establishment of standardized methods for detecting GM contents, and the development of rapid, high-throughput detection mechanisms for on-site real-world applications.
Identifying and defining the level of genetic modification in a particular variety
-
The molecular characteristics of GMOs include the entire inserted sequence of the exogenous gene, integration position, flanking sequences, and copy number. Molecular characteristic identification is the technical core for safety assessment. Prof. Zhang systematically used various techniques, including thermal asymmetric interlaced (TAIL)-PCR, genomic walking, and inverse PCR, to clone the integration sites and flanking sequences of GMOs that have been approved for import or independently developed in China, e.g., GM soybean (MON89788), GM rapeseed (T45, Oxy235), GM maize (MON863, TC1507, NK603, 3272, 59122), GM cotton (MON1445, MON531, MON88913, MON15985), and GM tomato (Huafan No. 1)[79−85]. He also developed formulae to quantify copy number of the exogenous gene based on quantitative real-time PCR (qPCR), and applied them to several GM crops[86−87].
In 2013, he proposed a new combination of second-generation sequencing technologies (TranSeq) for analyzing and identifying molecular characteristics at the whole-genome level[88]. By constructing paired-end (0.5–0.8 kb) and mate-pair (~10 kb) libraries and performing paired-end sequencing, the TranSeq algorithm and model could be used to comprehensively and accurately analyze molecular characteristics of GMOs[88]. He analyzed major GM rice varieties in China, such as TT51-1, T1C-19, T2A-1, RJ5, 114-7, G281, G6H1, and PA110, and found, for example, that the exogenous gene inserted on chromosome 10 of variety TT51-1, which has obtained a safety certificate in China, has undergone tandem duplication, and a new single-copy insertion has occurred on chromosome 4[88]. This information updating molecular characteristics data on GM crops has played an important role in obtaining renewed approval from the Chinese safety authorities and approval from the US Food and Drug Administration (FDA) and US Department of Agriculture.
Developing standardized methods for GM crop analysis
-
Since 1997, many countries such as the European Union, Japan, Korea, and China, have implemented a GMO labeling system. To ensure the smooth implementation of the labeling system in China, extensive research has been conducted on the qualitative and quantitative detection technologies of GM products by Prof. Zhang's group.
In 2004, he proposed defining an endogenous reference gene for GM crop detection. Target reference genes were chosen based on interspecies specificity (specific to a crop one species), intraspecies non-specificity (conserved between different varieties of a specific species), and stable low copy number. A strategy to identify and validate endogenous reference genes was established:
1. Use bioinformatics to analyze and screen candidate endogenous reference genes;
2. Design specific primers to amplify reference gene sequences in different cultivated varieties of the same species, and select genes with high sequence similarity;
3. Validate the candidate genes from the perspectives of interspecies specificity, intraspecies non-specificity, and copy number;
4. Establish qualitative and quantitative PCR assays based on the reference gene, and organize international collaborative experiments to evaluate the performance of the established assays.
Based on the above strategy, Prof. Zhang and his group successfully validated endogenous reference genes for rice SPS[89], canola HMG I/Y[90], tomato LAT52[91], cotton SadI[92], papaya Chy[93],and wheat gene[94].
These qualitative PCR and quantitative real-time PCR detection assays were used to monitor and detect the presence of the primary approved GM events in China, such as GM soybean (MON89788, MON87751, A2704-12, and A5547-127), GM canola (T45, Oxy235), GM maize (MON810, MIR604, MON863, TC1507, NK603, 3272, 59122), GM cotton (MON1445, MON531, MON88913, and MON15985), and GM papaya Huanong 1[79−85,93,95−98]. Prof. Zhang also organized international collaborative experiments to validate the developed methods, and demonstrate that these methods meet the requirements of qualitative and quantitative PCR methods for GMO detection[99−102]. These standardized methods have been successfully transformed into Chinese national standards and International Organization for Standardization (ISO) standards, and widely applied in GMO product detection in countries around the world.
In addition to establishing standardized methods for GMOs, he also placed a strong emphasis on developing certified reference materials (CRMs) for GMO analysis in China, which are under-resourced relative to the number of commercialized GM events. He initially proposed the concept of 'one crop, one reference material' and created novel plasmid and genomic DNA reference materials for GM soybean and maize[103−107]. These efforts resulted in the production of 14 plasmid DNA and genomic DNA reference materials covering 46 GM events used in China's commercial applications. Notably, rice TT51 (GBW10070), evaluated by his methods, was the first genetically modified 'first-grade certified reference material' in China. These new CRMs have successfully addressed the significant shortage of reference materials necessary for safety supervision in China, and relieved China's reliance on standards from the EU and USA.
Advancing rapid, accurate, and high throughput DNA detection methods
-
With the rapid development of commercialized GM crop events, quick screening and identification of GM events has become a challenge. Prof. Zhang developed several novel methods to improve/optimize nucleic acid extraction, target amplification, and identification of amplified products, such as Universal Template PCR (UT-PCR)[108]; Attached Universal Dual Probes PCR (AUDP-PCR)[109]; Microdroplet PCR Implemented Capillary Gel Electrophoresis (MPIC)[110]; and Visual Loop-mediated Isothermal Amplification (vLAMP)[111].
• In UT-PCR and AUDP-PCR, a common template sequence and primers are used to generate amplification products of different lengths for quantification purposes. These methods are applied to quantify gene copy number, pathogen load, transgenic composition, and microbes in environmental samples[108−109].
• MPIC combines the advantages of bipartite primers, microdroplet PCR, and capillary gel electrophoresis (CGE) for analysis multiple DNA targets. This method has good specificity and high sensitivity (0.1% for all targets), and has successfully amplified 24 different target DNA fragments from 14 GM events[110]. It is also flexible, high-throughput, and low cost due to multiple targets being detected in a single reaction. Moreover, preliminary results also indicate that MPIC can be used for semi-quantification.
• For vLAMP, he designed a simple device for extracting and visualizing DNA without using specialized laboratory instruments[111]. The simple DNA extraction device consists of a tailor-made filtration column containing silica gel membrane, and a modified medical syringe equipped with a sponge filter. Sample lysate filtration, DNA binding, washing, and elution occurs by applying air pressure using the syringe, thus eliminating the need for a centrifuge. The entire DNA extraction process can be finished within 10–15 min. The modified vLAMP method uses a microcrystalline wax–encapsulated bead in the reaction mixture, and fluorescent dye inside the bead can easily be released into the reaction mixture after LAMP amplification. The released fluorescent dye binds to the LAMP–amplified DNA and changes the color of the reaction mixture. This quick, simple and effective system can be applied successfully in on-field testing of GMOs, and vLAMP has now been adopted for onsite analysis of port inspection samples and practical field samples in China.
For the harmonization and standardization of GMO analysis methods globally, he constructed an open-access GMO Detection method Database (GMDD), which collects almost all previously developed and reported GMO detection methods grouped by different strategies (screen-, gene-, construct-, and event-specific)[112]. GMDD also provides user-friendly search services of the detection methods by event name, exogenous gene, or protein information, among others. Users can obtain the sequences of exogenous integration to facilitate PCR primer and probe design. Notably, the database archives contain various data on endogenous genes, certified reference materials, reference molecules, and the validation status of developed methods. To date, GMDD remains an informative database for GMO detection.
Introducing natural variation into the safety assessment of GMOs
-
Although the substantial-equivalent principle is widely believed to be applicable to the safety assessment of GMOs, puzzles about how to implement it in the real safety assessment exist. To make it work in reality, Prof Zhang employed the non-targeted metabolomics and introduced natural variations into the safety assessment of GMOs. He was the first person to profile the mature seed metabolomes of 100 rice cultivars[113], 29 common soybean cultivars[114], and 14 maize inbred lines[115], for such a purpose. He was also the first person to use such a strategy to assess the safety issues in GM maize event BVLA430101 over-expressing the Aspergillus niger phyA2[116], in which, he found that only levels of metabolites in the intended metabolic pathway are out of the range of the natural metabolic variations, providing a wonderful reference for GMO safety assessment.
Vale -
During his scientific research endeavors, Prof. Dabing Zhang fostered enduring partnerships with numerous colleagues, research institutions, and universities, both domestically and internationally. These concerted efforts have led Shanghai Jiao Tong University to establish dual-degree joint training agreements for undergraduate and graduate programs with both the University of Nottingham in the UK and the University of Adelaide in Australia. On January 23rd, 2015, Dabing's unwavering dedication culminated in the establishment of the collaborative UoA–SJTU Joint Laboratory on Plant Science and Breeding. This laboratory is situated in two locations: one at the Plant Research Centre, Waite Campus, at UoA, and the other at the Life Science Building, School of Life Sciences and Biotechnology, Minhang Campus, at SJTU. The primary objective of the Joint Lab is become a leading institution in cereal biology and breeding, exemplifying the exceptional collaboration between Australia and China. As a result of these collaborations and Dabing's tremendous research outputs, the international academic influence of SJTU in the field of plant science has experienced substantial growth. Additionally, his colleagues have gained access to an international cooperative platform, offering opportunities for their future academic pursuits. We extend our sincerest gratitude and respect for Dabing Zhang's invaluable contributions.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
Conflict of interest Rights and permissions (5) References(116) - About this article
Cite this articleYuan Z, Shi J, Yang L, Huang G, Liang W. 2023. Dabing Zhang (July 5, 1967–June 22, 2023). Seed Biology 2:11 doi: 10.48130/SeedBio-2023-0011 -
Dabing Zhang (July 5, 1967–June 22, 2023)
- Received: 09 July 2023
- Accepted: 11 July 2023
- Published online: 27 July 2023
We express our deepest condolences on the passing of Prof. Dabing Zhang, our esteemed mentor, colleague, and friend. Prof. Zhang, a member of the Editorial Board of Seed Biology, passed away at the age of 56. He served as a Chair Professor in the School of Life Sciences & Biotechnology at Shanghai Jiao Tong University (SJTU), a Professor at the University of Adelaide (UoA), and Director of the UoA–SJTU Joint Laboratory on Plant Science and Breeding. His research primarily focused on cereal development, including the identification of novel regulators governing reproductive architecture, male fertility, and responses to environmental cues such as temperature, photoperiod and mechanical signals, and genetically modified organism (GMO) biosafety assessment. Prof. Zhang leaves behind a dedicated staff, and numerous colleagues and friends.
-
Key words:
- Commemorative article /
- Dabing Zhang /
- Reproductive development













