-
Horizontal gene transfer (HGT), also known as lateral gene transfer, refers to the genetic information exchange between distantly related organisms and spreads across species boundaries, which is divergent from the typical vertical inheritance from parent to offspring. In nature, HGT does not occur equally among different domains of life. In prokaryotes, as one of the common evolutionary events, there was abundant evidence of HGTs in Eubacteria mainly mediated by plasmids[1]. HGT is central to the architecture and evolution of Eubacteria genomes and the recipient lineages benefit from HGT events by acquiring novel functions responsible for pathogenicity, bioremediation, xenobiotic tolerance, antibiotic resistance, metabolic detoxification, and so on[2−4]. HGT in Eubacteria is mainly realized by a three-step mechanism: transformation, conjugation, and transduction[5]. In contrast, HGT is less common in eukaryotes. In unicellular eukaryotes, it is generally believed that foreign genes enter into the recipient through symbiosis, infection, phagocytosis or other physical contacts, and then undergo homologous recombination and integration into the host chromosome. This hypothesis, put forward by Doolittle in 1998, is usually called gene transfer ratchet[6]. Compared to the relatively frequent HGT occurrence in unicellular eukaryotes, the relevant evidence is relatively rare in multicellular eukaryotes, and the shifting perception of HGT in multicellular eukaryotes occurred in 2007[7]. The authors showed that a large amount of DNA considered bacterial contamination in the previous sequencing was part of the insect nuclear genome, which confirmed the widespread HGT events from bacteria to insects. In particular, most of the observed HGT cases in eukaryotes have been derived from bacteria and fungi[8]. Albeit there are multiple reports, the underlying mechanism is still unclear, and several emerging evidence suggested that it is likely mediated by transposable elements (TEs) and other factors[8]. The most important characteristic of horizontal transposon transfer (HTT) different from HGTs is that TEs can move and amplify in the recipient genome, so they may be more easily transferred horizontally between different species. For example, a previous pioneering study about HTT has evidenced that the TEs underwent a horizontal transfer from Drosophila willistoni to D. melanogaster[9]. Although the relative scarcity of HGTs among eukaryotic animals, it has always aroused a particular interest and been identified in certain scenarios, especially in insects. Recently, the HGT events such as those identified from whiteflies and moths have conferred the insect lineages with stronger survival and reproductive fitness[10,11], albeit these cases have been documented in a relatively recent evolutionary event.
It is quite easy and comprehensible to confirm the occurrence of HGT events by characterizing the genes that are not supposed to be there. To date, plenty of examples have shown that many HGTs of insects have been derived from bacteria[10] (Fig. 1). Among these, the bacterial endosymbionts account for most HGT donors, and the host insects are capable of acquiring the genomes of bacterial endosymbionts by genetic integration, which was supported by the fact that Wolbachia and other endosymbionts colonized the host genomes of at least 20% of insect species[12]. In addition to the symbiont genomic integration via HGT, several examples have evidenced a single or few gene(s) transfers from bacteria, fungi, viruses and plants to insect genomes[10,13] (Fig.1). The horizontally transferred genes have been demonstrated to be implicated in the adaptation of recipient genomes[6]. However, due to the large variations in the genomic structure such as introns and GC content between insect lineages and candidate donors including bacteria, fungi, viruses, and plants, the recipients fail to take up foreign DNA upon most occasions and the newly acquired genes will be probably inactivated or eroded[14,15]. As a consequence, novel functional properties are unable to be generated in most HGT events from these distantly related donor species to insects. Despite these obstacles, recently an accumulating body of research has revealed that several insect HGTs are likely to become functional, such as detoxification, bacteriostasis, and promoting insect development[10] (Fig. 2). In this review, we provide an overview of the current advances in insect HGTs, aiming to integrate newly available examples associated with viruses, prokaryotes, and eukaryotes to insect transfers and discuss their nature of functionality (Table 1, Figs 1 & 2).
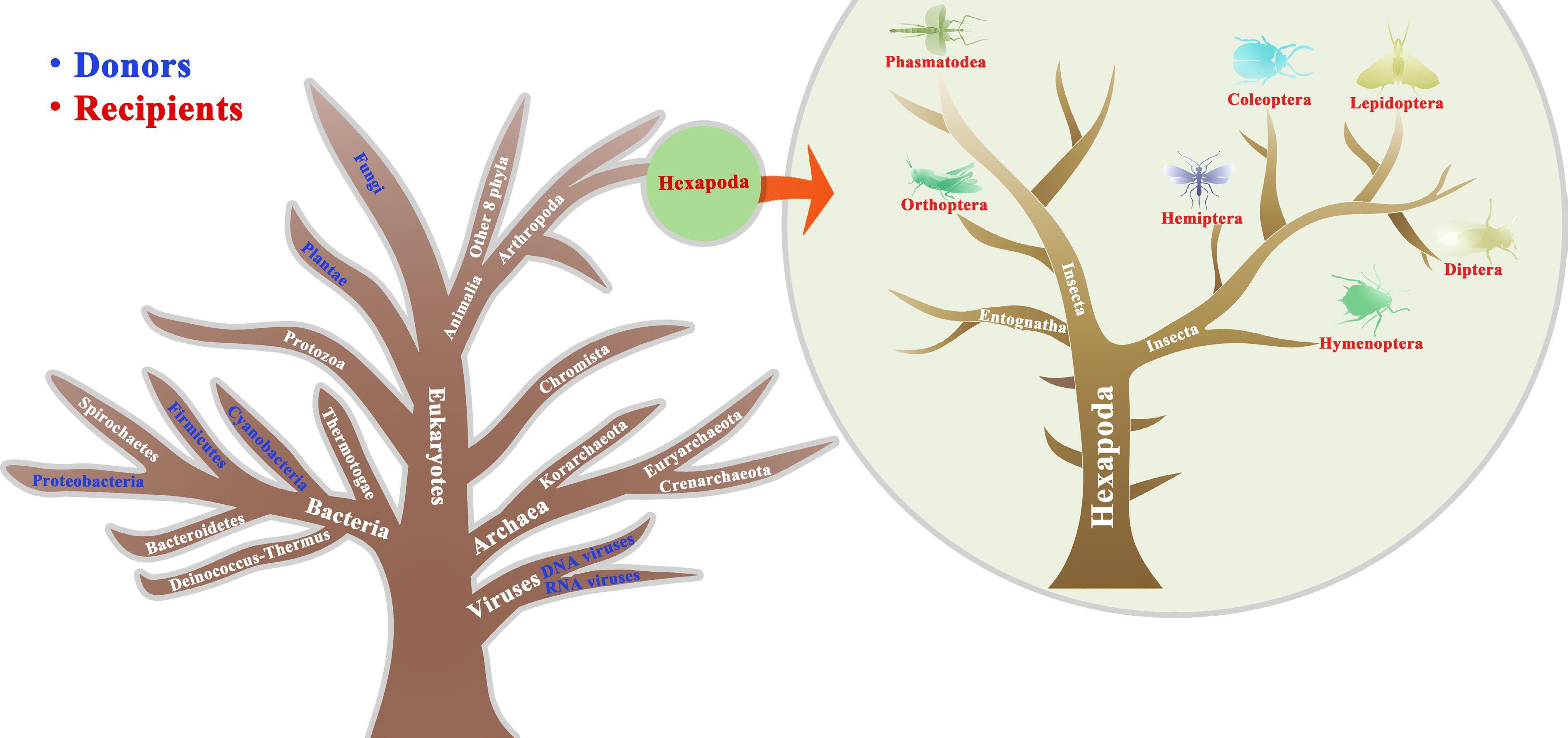
Figure 1.
Illustration of HGTs from other organisms to insects described in the main text. Blue and red represents the donors and recipients, respectively. The tree is designed only for illustration purposes merely explaining the donors and recipient insects that have undergone HGTs and is unable to accurately reflect a true phylogeny, which is modified from previous studies[1,4].

Figure 2.
An overview of HGTs in insects and mainly available examples associated with their functions. Part I: 'Donors' represent several groups providing the candidate horizontally transferred genes. 'Recipients' represent the nine insect groups that have received the horizontally transferred genes from donors. Part II: The molecular process of horizontal gene transfer. Part III: The most obvious examples of insect HGT events that have acquired novel functions.
Table 1. A list of horizontally transferred genes in insects.
Donor Recipient Gene name Functional category Reference Ascomycetes: undefined species Apriona japonica, Callosobruchus maculatus, Chrysomela tremula, Dendroctonus ponderosae, Diabrotica virgifera, Gastrophysa viridula, Leptinotarsa decemlineata, Phaedon cochleariae, Pissodes strobe, Sitophilus oryzae Coleoptera Pectin-degrading polygalacturonase (PG) Carbohydrate metabolism [72] Bacteria and fungi: undefined species Diabrotica virgifera Glycosyl hydrolase (GH45,
GH48, and GH28)[65] γ-proteobacteria: Cellvibrio, Teredinibacter Phaedon cochleariae Glycosyl hydrolase (GH11) [62] Bacilli: Bacillus Hypothenemus hampei Mannanase (HhMAN1) [13] Bacilli: Citrobacter, Enterobacter, Klebsiella Agrilus planipennis β-fructofuranosidase [64] Bacilli: Cedecea, Shigella, Yersinia Sphenophorus levis [63] α-proteobacteria: Wolbachia Callosobruchus chinensis wsp Undefined [25] 57 genes [36] Monochamus alternatus 31 genes [37] Zygomycetes: Blakeslea, Mucor, Phycomyces Asteromyia carbonifera, Chaitophorus populeti, Mayetiola destructor Diptera Phytoene
synthase and desaturaseFormation of body color [71] Cyanobacteria: Calothrix, Tolypothrix Aedes aegypti, Culex quinquefasciatus Ribosome inactivating gene Antimicrobic defense [68,69] Bacteriophage: APSE-2 Drosophila ananassae, D. biarmipes, D. bipectinata, D. primaeva, Myzus cerasi, M. persicae, Scaptomyza flava, S. nr. nigrita, S. pallida Cytolethal distending toxin B (cdtB) Defense against enemies [82] α-proteobacteria: Wolbachia Aedes aegypti, A. mascarensis, Culex pipiens quinquefasciatus AAEL004181, AAEL004188 Undefined [40] Drosophila ananassae nuwt [27] Glossina morsitans morsitans 16S rRNA, wsp, fbpA [26,41] Culex pipiens quinquefasciatus, Drosophila ananassae, D. sechellia, D. simulans Undefined [8,15] α-proteobacteria: Wolbachia; γ-proteobacteria: Buchnera Acyrthosiphon pisum Hemiptera ldcA, rlpA, AmiD, bLys, DnaE, AtpH, RlpA4 Amino acid metabolism, transport, and defense response [44, 45,
48−50]Saccharomycetes: Xanthophyllomyces; Zygomycetes: Phycomyces Carotenoid desaturase and synthase Formation of body color [32] Zygomycetes: Blakeslea, Mucor, Phycomyces Phytoene
synthase and desaturase[71] Plant: undefined species Bemisia tabaci Glucoside malonyltrasferase (BtPMaT) Phenolic glycoside detoxification [11] Ribosome inactivating gene Antimicrobic defense [75] β-proteobacteria: Candidatus; γ-proteobacteria: Carsonella Diaphorina citri ribC Riboflavin biosynthesis [51] γ-proteobacteria: Carsonella Pachypsylla venusta argH, cm, rsmJ, ribC, ydcJ Amino acid metabolism, transport, and defense response [52] α-proteobacteria: Wolbachia; γ-proteobacteria: Carsonella, Serratia, Sodalis Planococcus citri ≥ 22 genes Carbohydrate metabolism [54] Plant: undefined species Trialeurodes vaporariorum Ribosome inactivating gene Antimicrobic defense [75] Zygomycetes: Phycomyces, Rhizopus 34 Aphids Carotenoid desaturase and synthase Formation of body color [70] Chytridiomycetes: Rozella Copidosoma oridanum, Melittobia spp., Muscidifurax raptor, M. raptorellus, M. uniraptor, Nasonia longicornis, N. giraulti, N. Oneida, N. vitripennis, Spalangia cameroni, S. endius, Tachinaephagus zealandicus, Trichogramma pretiosum, Trichomalopsis sarcophagae, Urolepis rupes Hymenoptera Chitinase (GH19) Antifungal defense [73] α-proteobacteria: Wolbachia Nasonia giraulti, N. longicornis, N. vitripennis, Melittobia digitata Ankyrin repeat-containing gene Embryogenesis [8,38,39] Bracoviruses: Cotesia vestalis bracovirus Cotesia vestalis Helitron Undefined [80] Bacilli: Bacillus;
γ-proteobacteria: Cedecea, Rahnella, YersiniaAmyelois transitella, Bicyclus anynana, Bombyx mori, Danaus plexippus, Junoni coenia, Heliconius melpomene, Papilio glaucus, P. machaon, P. xuthus, Plodia interpunctella, Spodoptera frugiperda Lepidoptera β-fructofuranosidase Carbohydrate metabolism and alkaloid detoxification [56,61] Bacilli: Bacillus;
γ-proteobacteria: Serratia;
Baculovirus: Autographa californica nucleopolyhedrovirusBombyx mori Chitinase (GH19) Antifungal defense [55] Bacilli: Bacillus, Enterococcus, Lactococcus, Listeria, Streptococcus
α-proteobacteria: Methylobacterium, Wolbachia;
γ-proteobacteria: Edwardsiella, Photorhabdus, Providencia, Pseudomonas, Salmonella, Serratia;
Ascomycetes: Talaromyces24 genes Metabolic detoxification [58−60] Chlorophyceae: Chlamydomonas Dioxygenase Undefined [57] Bacilli: Bacillus, Enterococcus, Listeria, Streptococcus;
α-proteobacteria: Methylobacterium, Rickettsia;
γ-proteobacteria: Erwinia, Pseudomonas, Serratia, Yersinia;
Ascomycetes: TalaromycesDanaus plexippus 22 genes Carbohydrate metabolism and detoxification [59,60] Bacilli: Alicyclobacillus, Bacillus, Enterococcus, Lactobacillus, Listeria;
α-proteobacteria: Methylobacterium;
γ-proteobacteria: Pseudomonas, Serratia
Ascomycetes: TalaromycesHeliconius melpomene 20 genes [60] Ascovirus, baculovirus, poxvirus: undefined species Helicoverpa, Heliothis, Spodoptera Parasitoid killing factor (PKF) Defense against parasitoids [79] Bacilli: Enterococcus Plutella xylostella Glycosyl hydrolase (GH31) Metabolic detoxification [59] Bacilli: Listeria Alcohol dehydrogenase (LOC105383139) Courtship behavior [10] Bracoviruses: Cotesia kariyai bracovirus Spodoptera littoralis Gasmin Antimicrobic defense [78] α-proteobacteria: Wolbachia Chorthippus parallelus Orthoptera Minor capsid gene Undefined [28] γ-proteobacteria: Frateuria, Pantoea Aretaon asperrimus, Extatosoma tiaratum, Medauroidea extradentata, Peruphasma schultei, Ramulus artemis, Sipyloidea sipylus Phasmatodea Glycosyl hydrolase (pectinase) Carbohydrate metabolism [66,67] -
It is almost impossible to definitely determine an ancient HGT event occurring in an individual insect. The current gold standard for identifying an exogenous candidate as the horizontally transferred gene is molecular phylogenetic analysis. In addition, phylogenetic incongruence, patch distribution, and sequence similarity are good evidence, but in several cases, it is possible to provide a piece of more solid evidence by comparing the dS of the candidate horizontally transferred genes with the host genes. Meanwhile, codon bias and software are available to perform these analyses and draw a solid conclusion.
Phylogenetic evidence
-
To launch the program of HGT detection, a data set containing homologous protein or gene sequences of closely and more distantly related species in question should be first prepared followed by the phylogenetic construction. The incongruence within a well-established phylogeny can be used as the criterion to determine a HGT event[16]. In detail, if a gene of interest from the supposed species shows extraordinarily higher sequence similarities with its paralogs from distantly related species, thus strongly clustering into branches with at least 60% bootstrap, the HGT is considered reliable[17]. It is worth noting that there is a disparate detection pattern under different scenarios. Hereby, the nucleotide tree is more appropriate for studying a relatively recent HGT due to the larger variation in sequences, and the protein tree is better for detecting an ancient HGT event.
However, several potential drawbacks of this evolutionary inference have arisen and one of which is a lack of strongly supported evidence in phylogenetic reconstructions between the putative horizontally transferred genes of the candidates and these paralogues of the supposed donor species. For instance, the sequence with a short length is a case in point and such phylogenetic analysis based on these genes will generate an ambiguous tree with poorly supported bootstraps[16]. A similar situation possibly happens when an extraordinary tree emerges due to an unusual evolutionary rate or mode of the putative horizontally transferred genes[8]. This implies that a certain amount of HGT events are missed via the phylogenetic analysis. In contrast, some artifactual HGT events underlying strongly supported phylogenetic branches are likely to be falsely identified due to their different evolutionary selection pressures relative to native genes[8]. Moreover, it is difficult to distinguish a HGT event from an ancestral gene duplication or differential gene loss in diverse lineages under circumstances of inadequate taxon sampling[18−21]. Nonetheless, phylogenetic analysis is still considered the most compelling method to identify HGT events.
Genome-related evidence
-
Aside from phylogenetic analysis, another approach greatly relying on a wealth of sequenced genomic data can be applied to detect insect HGTs, and this detection tends to become more effortless with a higher assembly quality of many insect genomes throughout the last few years, thus contributing greatly to the recognition of ongoing HGTs[10,22]. A sense of the possible scope comes from the comparisons of gene structure, codon bias, and nucleotide base frequency in recipient genomes with those of closely related species[1]. This methodology has been initially developed to estimate the possible HGT events in bacteria[23,24], which has since been used in insect HGT detection. The optimal choice is probably the joint assembly of an assumed horizontally transferred gene with genes on a single genomic scaffold, in conjunction with obtaining active introns[14]. One of the most well-known cases was that the bean beetle X chromosome was embedded with at least a 11 kb fragment of DNA of the donor Wolbachia, leading to the premature termination of transcription and frame-shifting mutation of the coding regions of inserted DNA[25]. However, we should pay special attention to the symbionts or microbial contamination because of the difficulty in discriminating these interferences from very recent HGT events[26−28].
No remarkable difference in the codon usage pattern of coding DNA sequences has been observed within the genomes of intraspecific species, in contrast to the relatively large variations in interspecific species[16,29]. Thus, a HGT event can be alternatively detected by determining a different codon bias within the genomes of a range of candidate recipients. Furthermore, conversions in nucleotide base frequency between adjacent sequences, such as higher GC content, imply the insertion of a gene or non-coding region from candidate species into recipient genomes[1]. However, this appraising method can only identify relatively recent HGT events and fails to estimate an ancient HGT event due to the ameliorated nucleotide base composition and undistinguishable signal of exogenous segments in the recipient during a long-term convergent evolution[2,30,31].
Functional evidence
-
Transcription is necessary to function, and the straightforward evidence for determining the functionality of a horizontally transferred gene in insects is the transcription of mRNAs. To characterize the sequence and abundance of candidate transcripts, in situ hybridization has been extensively used. An excellent example was that a Wolbachia gene bound to a unique location on the host insect chromosome using fluorescence in situ hybridization (FISH) and Southern blotting[7,26,27]. However, the Wolbachia transcripts integrated into recipient genomes contained many pieces of pseudogenes, leading to the ambiguity of their function[4]. As a parallel measure, RNA-sequencing has also been used for detecting the transcription of possibly horizontally transferred genes, and the tissue-specific expression along with quantitative verification was likely to clarify their functionality[14,16]. Nevertheless, the gold standard for determining functional HGT is the phenotypic evidence correlated with expressions of proteins. Taking the pea aphid for instance, the horizontally transferred genes encoding enzymes for carotenoid biosynthesis were abundantly expressed in red individuals, thus playing a crucial role in the formation of body color[32]. Overall, these examples provide irrefutable evidence of the functional HGT events in insects.
-
Most exogenously prokaryotic DNA inserts into insect genomes can be divided into two scenarios: endosymbiotic and non-endosymbiotic origin. The endosymbionts to insect HGT is probably more common for the reason that endosymbionts are more constant and in close proximity to the cells of host insects[4]. A great deal of HGT events have been well-established, and suchlike transfers are of vital importance to the evolution of insects.
Wolbachia to insect HGTs
-
In the case of the vast majority, endosymbionts to insect HGT seems to be originated from Wolbachia as yet, a member of Rickettsiales belonging to α-proteobacteria. Wolbachia has colonized various insect species (at least 20%), such as Coleoptera, Diptera, Hymenoptera, and Hemiptera[33,34]. The maternal inheritance of Wolbachia via cytoplasm created an excellent opportunity to integrate candidate genes into the genomes of host insects, thus acting as an ideal donor for HGT[35]. An engaging discussion about the maintenance of HGT events from Wolbachia to certain insect species has always been addressed. We now turn to the cases of HGT events from endosymbiotic Wolbachia origin to recipient insects.
The first example of Wolbachia to insect HGT was described in detail in the adzuki bean beetle Callosobruchus chinensis, where a genome fragment including a gene encoding Wolbachia surface protein (wsp) was transferred into the X chromosome of the recipient insect[25]. This is the early experimental evidence illustrating the HGT event between Wolbachia and the host insect. Subsequently, investigations through PCR detection and Southern blot confirmed that 57 genes derived from Wolbachia were integrated into the C. chinensis nuclear genome, albeit the transcriptional levels of ~50% genes were relatively low followed by being evidenced using FISH analysis[36]. A parallel work based on exhaustive PCR surveys and FISH analysis determined that 31 of 214 Wolbachia genes were transferred and located on an autosome of two populations of Cerambycidae Monochamus alternatus[37]. Besides, in Chorthippus parallelus, it was also found that at least 448 and 144 kb of DNA fragments from two discrete Wolbachia supergroups, B and F, were integrated into the nuclear genome of host grasshoppers, and FISH indicated endosymbiotic genome inserts in host chromosomes[28]. Remarkably, it was the first case of scanty HGT events discovered in orthopteran insects.
A survey conducted by Hotopp et al. comprehensively examined 20 insect genomes for potential endosymbiont Wolbachia to insect HGTs and provided credible evidence of such transfers in dipteran and hymenopteran insects, including D. ananassae, Nasonia giraulti, N. longicornis, and N. vitripennis by combining PCR, high-throughput sequencing and fluorescence microscopy analyses. However, the inserted fragments ranged from nearly the entire Wolbachia genome (> 1 Mb) to short fragments (< 500 bp) and some of the horizontally transferred genes were transcribed within cells of antibiotic-cured insects, confirming the occurrence of insect heritable HGT events[7]. Subsequently, genomic and evolutionary analyses performed by Werren et al. demonstrated that three Nasonia lineages, N. giraulti, N. longicornis, and N. vitripennis, acquired one or more laterally transferred genes encoding ankyrin repeat-bearing proteins from Wolbachia origins by duplication and divergence, their transcriptions during various stages of both sexes were verified by EST or tiling microarray analyses[38]. A more recent example of the HGT event from endosymbiotic Wolbachia to insects also comes from the wasp superfamily Chalcidoidea, it has been revealed that a large family of ankyrin domain-encoding genes originated from Wolbachia underwent a complicated evolutionary history with multiple instances of HGTs, and these transferred genes were ubiquitously expressed throughout the embryos acting in patterning, morphogenetic movements, and relative timing of N. vitripennis and Melittobia digitata embryonic events[39].
In dipteran insects, take two Aedes species, A. aegypti and A. mascarensis, for instance, the HGT events, involving two adjacent genes and their homologs, were respectively identified from Wolbachia symbiotic bacteria origins, and the expression of these genes was analyzed by qPCR and microarray[40]. Similarly, Doudoumis et al. performed a series of PCR detection using 16S rRNA, wsp, and fbpA gene markers to identify potential HGT events, in which genes of Wobachia origins were inserted into the nuclear genome of tsetse fly Glossina morsitans morsitans[41]. A further study from Brelsfoard et al. validated the two large insertions of Wolbachia DNA in the genome of G. morsitans morsitans, and southern blot combined with FISH analysis revealed that these insertions located on the autosome and sex chromosomes, however, most horizontally transferred genes present in the insertions were unable to be transcribed[26]. Furthermore, on the basis of re-sequenced genomes of three D. ananassae lines, the copy numbers of Wolbachia DNA transferred to the host nuclear genomes were determined followed by the detection on host chromosomal localization using FISH analysis, and the result revealed that different parts of these HGTs varied a lot in different lines, suggesting their varying degrees of evolutionary selection pressures[27]. A follow-up study conducted by Choi et al. systematically characterized the integrated fragments of Wolbachia colonizing D. ananassae using genome sequencing involving 15 strains, and at least two copies were observed in most of the integrated regions, implying widespread double or duplicated integration of Wolbachia DNA in dipteran recipient genomes[15].
Other endosymbionts to insect HGTs
-
As a commonly symbiotic partner within host insects, Wolbachia endows hosts with favorable effects[34]. In parallel, the mutually endosymbiotic relationships underlying the aforementioned HGT events have also been highlighted between various bacterial lineages other than Wolbachia and host insects[42,43]. Here we provide extra examples to introduce the endosymbionts to insect HGTs other than Wolbachia, thus deeply advancing our understanding of the adaptive evolution between endosymbionts and host species.
It was first discovered that aphids harbor an obligate mutualist, Buchnera aphidicola (γ-proteobacteria), and encoded genes from this heritably bacterial origin[43,44], assuredly supporting the horizontal transfer of ancestral symbiont genes to insects. Several lines of evidence including Southern blot and qPCR analysis confirmed the presence of these genes in the aphid genome along with their high expression levels in the bacteriocyte. Similarly, it was further demonstrated that aphids acquired these laterally transferred genes, ldcA and rlpA, from a rickettsial bacterium Buchnera, closely related to Wolbachia, responsible for the provision of nutrients[45]. Previously functional investigations based on these genes indicated that ldcA encodes a carboxypeptidase essential for the recycling of cell wall polymer murein[46], while rlpA encoding a lipoprotein worked in tandem to degrade peptidoglycan[47]. The ongoing investigations via whole genome sequencing, phylogenetic and experimental analyses suggested that the pea aphid Acyrthosiphon pisum acquired 12 genes or gene fragments including three ldcAs, five rlpAs and four other metabolic genes from the genome of its symbionts Buchnera, suggesting a set of duplicated events of the transferred genes in the context of aphid-Buchnera symbiosis, and among these, at least eight genes appeared to be responsible for the synthesis of numerous essential amino acids which were highly expressed in bacteriocytes[48,49]. The intimate symbioses between bacterial lineages and host insects have repeatedly evolved, and Nakabachi et al. reported the first case of organellogenesis from the endosymbiotic origin in symbiont-insect systems. It was found that a horizontally acquired gene of aphid A. pisum encoding a protein RlpA4 was derived from a bacterium other than Buchnera and the specific expression in the maternal bacteriocyte was analyzed and verified by immunoblot and immunomicroscopy, which was transported into an obligate endosymbiont Buchnera[50].
Apart from aphids, the Asian citrus psyllid Diaphorina citri harbors two distinct obligate symbionts, a nutritional symbiont Carsonella (γ-proteobacteria) and a defensive symbiont Profftella armatura (β-proteobacteria)[51]. The genomes of symbiotic Profftella lost a ribC gene required for riboflavin biosynthesis that was horizontally acquired by host D. citri, reflecting an ancient HGT event and their mutual genome communication for functional complementarity[51]. To disclose the parallel histories of such HGTs in other sap-feeding psyllids also harboring a symbiont Carsonella, the gene contents along with their expression patterns of Carsonella were analyzed in host Pachypsylla venusta and a remarkably similar set of laterally transferred genes integrated into the host genome were detected[52]. Phylogenetic analysis placed these genes with the orthologs of Carsonella origins into a monophyletic group, strongly supporting that Carsonella acted as a donor of HGT. Also, several acquired genes appeared to be derived from groups with very common endosymbionts, including Rickettsia and Wolbachia, suggesting that these candidates were from multiple donor lineages[52]. In a previous study, most of these genes were dramatically transcribed in psyllid bacteriomes housing Carsonella ruddii, possibly making up for the gene losses in the endosymbiont genome, and further functional inferring based on metabolic pathway annotation and qPCR analysis indicated that these transferred genes were required for the biosynthesis of arginine and phenylalanine pathways[53]. In contrast, a study implicated in bacterial genome sequencing and phylogenetic analyses detected that at least 22 genes of the mealybug Planococcus citri had histories of horizontal transfers from diversely facultative symbiont origins, mainly including Wolbachia, Sodalis, and Serratia[54]. Analogous to psyllid recipients, the ongoing transcriptome analysis recorded greater expression values of these horizontally transferred genes in mealybug bacteriomes as well, and a strikingly similar range of functions contributing to the biosynthesis of essential amino acids, vitamins, and biotin were shaped by independent HGT events from endosymbiont origins to two hemipteran insects[54]. Taken together, these cases are living representations of the HGT events crucial for driving the co-evolution between bacterial mutualists and their host insects.
Other prokaryotes to insect HGTs
-
In addition to the widely known HGTs from endosymbionts to insects, there is a growing body of literature that recognizes the importance of HGTs sourced from non-symbiotic bacteria conferring great benefits on insect recipients. As exemplified in a representative insect of Lepidoptera, Bombyx mori, sequence alignments combined with phylogenetic analyses placed the genes encoding chitinase and β-fructofuranosidase with the bacterial paralogs, respectively, derived from Serratia and Bacillus, into one cluster[55,56]. The location of chitinase on silkworm chromosome 7 was confirmed by Southern blotting followed by the stage- and tissue-specific expression profile examinations using Northern blotting, implying its potential antifungal activity[55]. Simultaneously, the accumulation of β-fructofuranosidase transcripts in silkworm midgut was shown, and the protein immunofluorescence localization within the midgut goblet cell cavities along with the determination of enzymatic activity substantially demonstrated its unique attribute of detoxifying alkaloids highly toxic to insects that are unlikely to feed on mulberry leaves[56]. The next genome-wide screening combining phylogenetic analysis defined most horizontally transferred genes in B. mori as entomopathogenic bacterial origins, such as Bacillus, Enterococcus, Lactobacillus, Salmonella, and Serratia, and further biochemical pathway prediction based on EST checking and microarray expression signals categorized these genes into several functional groups indispensable for physiological processes, especially in metabolic detoxification[57,58]. Follow-up comprehensive supplementary analyses involving four insect genomes, including B. mori, Danaus plexippus, Heliconius melpomene, and Plutella xylostella provided solid evidence of the occurred HGT events within lepidopterans, which supposedly participated in xenobiotic metabolism[59,60].
The duplication or loss events of horizontally acquired genes seem to have independently occurred in different insect species, thus widespread in lepidopteran but sporadically occurring in coleopteran and hymenopteran, possibly to better adapt to the recipient genomes[61]. On this basis, Dai et al. performed a comprehensive evolutionary analysis of β-fructofuranosidase genes acquired from bacterial origins in two lepidopteran insects, B. mori and Papilio xuthus, and found that these genes embedded in different insect recipients were all highly expressed in larval silk gland and midgut. There was a marked divergence in the enzymatic properties of β-fructofuranosidase in breaking down sucrose mortierellate, and the loss of function mutant generated by CRISPR/Cas9 displayed a delayed insect development and an impaired ability to detoxify alkaloids, indicating its functional diversification including metabolic and digestive adaptation[61]. Recently, a breakthrough dedicated to the comprehensive screening of HGT events in 218 insects had been made in which the authors found that these insects laterally acquired more than a thousand genes with a diversity of functions from prokaryote donors via 741 disparate HGT events[10]. It was especially noteworthy that the average highest acquired gene numbers were recorded in lepidopteran recipients, and further in-depth functional excavation based on the prevalent HGT-acquired alcohol dehydrogenase gene (LOC105383139) in diamondback moths, P. xylostella, indicated that the acquired gene from a bacterial donor Listeria resides in the autosomes and contributed to male courtship behavior[10]. Altogether, these horizontally acquired genes embedded in the genomes of moths and butterflies provide a novel insight into their potential contributions to ecological adaptability in recipient insects.
In coleopterans, a previously non-reported bacterial gene (HhMAN1) encoding a typical glycosyl hydrolase, mannanase, hydrolyzing coffee berry galactomannan was first identified in the genome of coffee berry borer beetle Hypothenemus hampei, which was species-specific and unable to be detected in genetically close species, and its universal presence in individuals of a broad geographic collection spanning 16 countries indicated an ancient HGT event[13]. As one of the prevalent donors of adaptive genetic materials, bacterial genes have continually been found in many other coleopteran lineages. In mustard leaf beetle, genes from bacterial origins, glycosyl hydrolases (xylanases) were also found in the genome of Phaedon cochleariae, further evolutionary evidence and enzymatic activity analysis suggested that P. cochleariae originally acquired these genes from γ-proteobacteria Cellvibrio and Teredinibacter required for degrading xylan through the best-known mechanism, HGT[62]. Also, take emerald ash borer Agrilus planipennis and sugarcane weevil Sphenophorus levis for examples, genes encoding β-fructofuranosidase were characterized in two insect genomes and qPCR analyses revealed their peak expressions in digestive apparatus and midgut, and phylogenetic analyses indicated that the β-fructofuranosidase of A. planipennis and S. levis were closely similar to the paralogs of Firmicutes groups, such as Citrobacter, Enterobacter and Klebsiella, suggesting the bona fide cases of HGTs in beetles[63,64]. Further analysis combining enzyme assays and chromatography indicated the β-fructofuranosidase of A. planipennis could hydrolyze raffinose and sucrose[63]. Likewise, next-generation sequencing and assembled transcriptomes of western corn rootworm, Diabrotica virgifera, indicated the exclusive presence of three GH family genes (GH45, GH48 and GH28) in Chrysomeloidea and Curculionoidea superfamilies, demonstrating their lateral acquisitions from candidate donors and probably representing an adaptation to a specific ecological niche for phytophagous beetles[65].
More particularly, six diverse stick insects belonging to Phasmatodea horizontally acquired the glycosyl hydrolase encoding genes, pectinases, from bacteria, which were highly expressed in the anterior midgut based on transcriptomic data analysis[66]. A more recent study conducted by Shelomi et al. also clarified the evolutionary origins of the horizontally acquired pectinases in stick insects and found that the donors of these transferred genes can be traced to Proteobacteria[67]. It is quite reasonable to assume that these genes have contributed to degrading plant cell walls into monomer components and increasing digestive efficiency. In mosquitoes, Lapadula and colleagues recently reported definite evidence of the horizontal transfers of ribosome inactivating genes in A. aegypti and Culex quinquefasciatus and experimentally confirmed their transcriptions in different developmental stages. To unveil their phylogenies, an integrated study combing taxonomic distribution evaluation and phylogenetic inferences was conducted, supporting that mosquitoes acquired these genes from a cyanobacterial donor species via a HGT event[68,69]. However, there are still many unanswered questions about the potential physiological functions of these horizontally transferred genes. Given all evidence mentioned so far, one may suppose that the prokaryote to insect HGT is a potentially critical source of genetic materials for shaping novel functional characteristics in lineage-specific recipients.
-
The continuing tracking of diverse gene origins and a large number of complicated phylogenies have led to a solid conclusion that many genes have indeed undergone horizontal transfers from eukaryotic origins to insects, albeit it has been highly underappreciated during the past decades. However, several pieces of evidence have re-attracted much attention recently[10,11,22]. Of burgeoning significance is the accumulation of cases describing HGT events from fungi and plants to insect lineages, which contributes to figuring out their functional properties.
Fungi to insect HGTs
-
To date, several genes derived from fungi have been transferred to insect recipients, and the earliest case of such transfers has been elaborately described strikingly in aphids. It is widely known that the carotenoids biosynthesis pathway is absent in arthropods including insects, but indispensable for various physiological functions such as pigmentation. Moran & Jarvik found that multiple genes encoding carotenoid desaturases and synthases were integrated into the genome of pea aphids, which were required for the carotenoid biosynthesis in A. pisum. Further evolutionary analysis indicated that these genes discovered in aphid genome underwent horizontal transfers from fungal origins along with gene duplications, and the generation of desaturase deficient mutants resulted in the loss of torulene in aphids, thus displaying a phase transition from red body to green[32]. Subsequently, a more elaborately comparative analysis based on sequences of 34 highly diverging aphid lineages together with fungal sequences retrieved from databases supported a shared origin of the horizontally transferred desaturases from fungal species of Mucoromycotina and their copy numbers varied widely from one copy to seven copies in different aphid species, obviously diverse from the consistently single copy in fungal genomes[70]. These acquired genes of fungal origins combined with their expression profiling were in good accordance with the carotenoid biosynthesis in different aphid lineages.
In dipteran insects, the carotenoid-related genes residing in the genomes of gall midges were also detected, which were homologous to fungal genes, and further phylogenetic analysis showed that these homologs from fungi lineages were laterally transferred to insect recipients[71]. As a pivotal innovation, the cross-species HGTs associated with carotenoid-related genes may account for the strong environmental adaptability of phytophagous insects and facilitate their extensive diversity across plant lineages. Sporadic occurrences of HGT events from fungi origins were also discovered in a wide range of lepidopteran species including B. mori, D. plexippus, and H. melpomene, and a transferred gene encoding dioxygenase was simultaneously identified in three recipient species, implying ancient HGT events[60]. One more particularly fascinating case comes from herbivorous beetles, on the basis of an increasing number of genomes of leaf beetles and weevils in which the genes encoding pectin-degrading polygalacturonases were embedded, Kirsch et al. subtly analyzed the evolutionary origins of polygalacturonases using the transcriptome data of 10 beetles by comparing with their counterparts including bacteria, fungi, and plants, demonstrating that these genes shared a common ancestor, the ascomycetous fungi. Further heterologous expressions of polygalacturonases and enzymatic activity assays indicated these orthologues were a set of lineage-specific enzymes in degrading pectins[72]. It is notable that a previously recognized source of parasitoid genome innovation has also turned out to be the HGT event. In detail, the genomes of 15 Chalcidoidea wasp species respectively encoded a chitinase venom gene (GH19), and phylogenetic analysis placed these genes with paralogs derived from Rozella allomycis into one clade, indicating an occurrence of the HGT event[73]. A remarkable accumulation of mRNAs was further recorded in the venom glands of half of the species harboring this gene, and in-depth RNAi analysis in the model wasp N. vitripennis disclosed that it incorporated novel function involved in antifungal defense into venom repertoire, most probably mapping to other insect lineages. To sum up, these inevitable HGT events traced from fungal footprints greatly promote the evolution of insect recipients to maximize their fitness.
Plants to insect HGTs
-
As an unconsciously emerging field of studies, rare cases of plants to insect HGTs have been previously reported, there are however many gaps which need to be filled in this research. Unlike the relatively easy detection of prokaryotes to insect transfers, such events are indistinguishable due to the more similar genetic backgrounds between plants and herbivorous insects. Although there are no large-scale plant contributions documented in genomes of insect lineages, several potentially interesting cases emerged in silkworms and whiteflies[57,74]. In particular, Zhu et al. defined a candidate dioxygenase gene in B. mori as a horizontal transfer from a plant donor combining comprehensive genome sequence alignment and phylogenetic analysis. From the co-expression data mapped onto a biological pathway, it was speculated that this acquired gene was implicated with the biosynthesis of antibiotic products, potentially endowing its resistance to pathogens[57].
Several recent HGT events that have otherwise been found in insects lie in the whitefly, and the gene transfers from plant origins formed the central focus of a study in which Lapadula and colleagues found that ribosome inactivating genes integrated into the genomes of Bemisia tabaci and Trialeurodes vaporariorum were independently ancient HGTs from plant donors, and RNA-sequencing evidence demonstrated their transcription and splicing, suggesting the functionality in environmental adaptation[75]. In particular, plant lineages produce phenolic glucosides, a conventional class of secondary metabolites, to poison phytophagous insects and herbivores have evolved multiple countermeasures to modify and detoxify these plant-derived toxicants[76,77]. A recent ground-breaking investigation in which a novel function was acquired by B. tabaci has been elaborately interpreted[11]. The author found clear evidence that B. tabaci hijacked a plant lineage-specific gene (BtPMaT1) encoding glucoside malonyltrasferase, certainly traced to a plant origin via a HGT event to successfully neutralize plant toxins based on phylogenetic construction combined with metabolic profiling and gene knockdown analysis[11]. An additional gene copy, BtPMaT2, was also identified by Xia et al.[11] but the authors failed to reveal its function. These plants to insect HGT events open up new horizons about how generalist herbivores commandeer the defensive weapon of plant lineages, contributing to unraveling the molecular basis of plant-herbivore interactions[74].
-
To date, scarce gene transfers have been demonstrated within viruses-insect relationships, albeit it has been around 20 years since the first case of the virus to insect HGT was identified[55]. Here, we gather strong indications lying in HGTs from virus donors to insect recipients rather than being other instances of prokaryotes and eukaryotes to insect HGT events, which raises the question of how these acquired genes shape the orientation of insect evolution.
A pioneering exploration into such area comes from the case of the silkworm in which a novel chitinase gene sharing an extensive similarity with the sequence of an ancestral baculovirus, Autographa californica nucleopolyhedrovirus, was identified as the horizontally transferred gene, and its expressions in epidermis and midgut were specifically profiled during B. mori ecdysis and pupation[55]. Worth a mention, is that in nature, HGTs from symbiotic viruses of parasitoids to host insects also occur in lepidopterans possibly because their larvae and pupae are frequently attacked by wasps. Di Lelio
et al. reported that a gasmin gene ancestrally residing in the genome of the endosymbiotic virus of braconids was transferred to the donor species Spodoptera littoralis, and a higher expression level of gasmin was recorded in larval circulating hemocytes, where a battery of transcripts rapidly generated after immune priming of pathogens. Further phenotype characterization based on dsRNA injection disclosed its role in facilitating hemocyte phagocytosis to withstand the invading bacteria[78]. A recent paralleled study also implied the potential prevalence of HGT events between viruses and lepidopteran lineages[79]. In this study, Gasmi et al. characterized a protein family, parasitoid killing factor (PKF), extensively present in ascovirus, baculovirus, entomopoxvirus, and lepidopteran hosts including Helicoverpa, Heliothis and Spodoptera species, and the occurrence of HGT events was substantially confirmed by evolutionary construction among these species. The authors further provided several pieces of evidence that the PKFs derived from S. exigua and nucleopolyhedrovirus were specifically toxic to parasitoids in Microgastrinae subfamily through a mechanism of inducing cellular apoptosis and eliciting DNA fragmentation in susceptible parasitoids[79]. Similarly, Heringer & Kuhn described a multilevel HGT event in which the gene Helitron of endosymbiotic bracovirus origin invaded the genome of host Cotesia vestalis followed by a later horizontal transfer from wasp to the lepidopteran host[80]. Based on molecular evolutionary analysis of various geographical populations, the authors pointed out that both the bracovirus to wasp and wasp to lepidopteran HGTs occurred in East Asia populations. Remember that the DNA fragments of polydanvirus residing in the calyx of parasitoids are transmitted into lepidopteran hosts during parasitization[81], we argue that this parasitic behavior contributes to such HGTs because of the accessible genetic resources for candidate recipients. In brief, these results demonstrate that the key physiological characteristics of lepidopterans do not necessarily derive from a straightforward evolutionary event and alternative benefits can be acquired from mutualistic viruses of their natural enemies via a roundabout HGT event, highlighting the evolutionary arm race in virus-parasitoid-lepidopteran tritrophic interactions. Another highlight about the physiological traits conferred by such analogously accidental viruses to insect HGTs comes from aphids and fruit flies. The authors manifested that a gene, cytolethal distending toxin B, was integrated into nuclear genomes of aphids and drosophilid species via an initial HGT event in which the bacteriophage infecting Candidatus Hamiltonella defensa certainly served as one of the alternative donors followed by later interspecific horizontal transfers from an ancestral Myzus spp. to Drosophila subgroups and an interordinal D. ananassae to D. biarmipes transfer, which showed a higher expression in Drosophila larvae and retained enzymatic activity to degrade DNA[82]. It is deduced that these domesticated genes have undergone an ancient horizontal transfer from bacteriophage ancestors to diverse insect species and are likely to confer insect lineages’ resistance for defending against natural enemies.
-
In this review, both incipient and recent HGT events have been heatedly discussed in insect lineages, uncovering the truth beneath the mask that the genes have been transferred from the plausibly related donors spanning viruses, bacteria, fungi and plants to recipient insects. Among these, a plethora of horizontally transferred genes have been derived from prokaryotic lineages, especially the endosymbiont origins. It is unquestionable that the HGT randomly occurs at a relatively low frequency in taxonomic insect lineages and the majority appear to be nonfunctional which will be eroded during the long-term convergent evolution between insect recipients and donor species. However, there are several clear examples that the horizontally transferred genes in insects are indeed transcribed in stage- and tissue-specific manners and particular interest has always been raised in their roles in maintaining and acquiring novel functions such as development and reproduction regulation, environmental adaptation, and immune defense.
Albeit progress in the insect HGT field has been remarkable over the past few years, and these findings have gone some ways toward enhancing our understanding of the evolutionary history of insect lineages, many questions and deficiencies in the current research have been identified as being in need of further investigation. Of these, the most notable one is that there is a lack of rigorous and systematic methods to identify the horizontally transferred genes, and the donor groups have not been fully considered. Eventually, it is necessary to put forward a simple and feasible solution. Additionally, for more closely sourced metazoan donor species, such as the natural enemies, preys, and competitive species, more accurate methods need to be established. Considering this point, the genomes of many insect species along with their closely related relatives are in urgent need of being advanced and the recently flourishing third-generation single molecule sequencing will provide us with a novel insight into establishing a greater degree of accuracy on such HGT events. Secondly, the current study of HGTs in insects is largely concentrated in the groups of Coleoptera, Diptera, Hemiptera, Hymenoptera, and Lepidoptera, which is relatively limited to phytophagous and parasitic insects. The interaction between plant donors and recipient insects is an important driving force for the speciation and species diversity of insects. HGTs between plants and insects should be given further attention. On the other hand, to fully uncover the mystery of HGTs in insects, it is necessary to explore more recipient groups, such as the predatory, saprophagous, and fungivorous insects. Far more important is that the functional evidence of insect HGTs is scarce and the functional verification of horizontally transferred genes is extremely difficult. Looking back at the history of our knowledge, future studies should be addressed to express these newly acquired genes and obtain their additionally functional scopes, making the complex evolutionary history of intraspecific and interspecific species more accessible.
The authors acknowledge the Hainan Province Science and Technology Special Fund (ZDYF2023XDNY075 & ZDYF2021XDNY302), the Key Project of National Natural Science Foundation of China (31830074), the Hainan Yazhou Bay Seed Laboratory Fund (B21HJ0401) and the funding of Hainan University (RZ2100003220).
-
The authors declare that they have no conflict of interest.
-
Received 30 November 2022; Accepted 2 March 2023; Published online 17 March 2023
-
# These authors contributed equally: Binglin Xing, Lei Yang
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xing B, Yang L, Gulinuer A, Ye G. 2023. Research progress on horizontal gene transfer and its functions in insects. Tropical Plants 2:3 doi: 10.48130/TP-2023-0003
Research progress on horizontal gene transfer and its functions in insects
- Received: 30 November 2022
- Accepted: 02 March 2023
- Published online: 17 March 2023
Abstract: Horizontal gene transfer (HGT) refers to the asexual exchange of genetic information between distantly related organisms. Although it is well acknowledged that HGT greatly contributes to the adaptive evolution in prokaryotes, its significance in shaping the orientation of eukaryote evolution remains obscure, especially in insect lineages. The massively expanded genomic data appears to be an excellent choice to uncover the mystery of HGTs in insects nowadays. Here we gather a body of evidence showing the HGT events from three broad donor origins, viruses, prokaryotes, and eukaryotes, in which most horizontally transferred genes are unlikely to be functional and will be eroded as a result of the difference of inheritable background between insect recipients and donor species. Nevertheless, particular interests in the prominent role of insect HGTs in maintaining and acquiring new functionalities have still been raised to underpin their adaptations. Among these, the previously investigated properties including reproduction regulation, detoxification of plant metabolites, formation of body color, and antimicrobic immunity are mainly included in different insect lineages. Albeit such cases are just the tip of the iceberg, we demonstrate that HGT drives insect evolution, especially in coevolution with host plants, and additional explorations into its functions should be given attention in order to access the complex evolutionary history of insects in the near future.
-
Key words:
- Horizontal gene transfer /
- Adaptive function /
- Recipient insect /
- Donor












