-
For forage and turf grass species, regulation of leaf senescence has a significant impact on both the biomass yield and quality of forages and the amenity of turfgrasses. Preventing or delaying precocious leaf senescence is important to improve forage yield, enhance forage quality, and maintain a long-term green lawn and turf. Leaves are the primary photosynthetic organs sustaining the energy supply in plants. Senescence is the final stage of leaf growth and development. It involves the degradation of chlorophyll (Chl), carbohydrates, fats, proteins, and nucleic acids and the remobilization of organic and inorganic nutrients to newly developed tissues and storage organs within the same plant[1]. In agricultural production, precocious leaf senescence causes a decrease in crop photosynthetic efficiency and yield[2,3].
Multiple endogenous and external factors influence the progression of leaf senescence, such as plant and leaf age, growth and development, plant hormones, light, drought, salt stress, pathogen infection, and herbivore attacks. When the environment is unfavorable for plants, nutrients are transferred from old leaves to new leaves, seeds, and other sink organs (e.g., stolons/rhizomes/shoot crowns) to sustain their survival. Therefore, the progression of leaf senescence is rarely at a constant speed but rather a complex and ever-changing process. The occurrence of various interactions among the developmental processes of leaf senescence, growth of the whole plant, plant reproduction, and environmental adaptation underscores the importance of leaf senescence in the overall life cycle of plants[4].
Color changes in leaves are the most remarkable sign of leaf senescence. With the gradual degradation of Chl and accumulation of carotenoids and anthocyanins, leaf color transitions from green to yellow or red. In the early stages of leaf senescence, the chloroplast membrane is damaged; however, the chloroplast size remains unchanged, whereas in the later stages, chloroplasts rapidly decrease in size, and the thylakoid membranes disintegrate. Through intracellular autophagy, the damaged chloroplasts collapse and are then transported to vacuoles for further breakdown by hydrolases[5]. As the cells progress from senescence to death, they still require energy to sustain their activities. In the early stages of senescence, cellular energy depends on the mitochondrial supply because of Chl degradation and reduced photosynthesis. Therefore, mitochondria remain intact during the later stages of senescence, thereby maintaining their ability to provide energy to cells. In leaves, up to 70% of the total nitrogen content are in chloroplasts. During leaf senescence, proteins in chloroplasts degrade to amino acids that are redistributed to other plant organs to enhance nitrogen use efficiency[6]. The selective catabolism of amino and fatty acids supplements α-ketoglutarate and glutamate in cells, providing a carbon chain for nitrogen remobilization, which facilitates nitrogen transport in aging leaves[7].
From the perspective of molecular genetics, leaf senescence involves chromatin remodeling, DNA transcription, and post-transcriptional, translational, and post-translational fine-tuning mechanisms. For example, the regulation of chromatin encompasses nucleosome assembly, disassembly, and rearrangement, DNA methylation, and post-translational covalent modifications of histones (including acetylation, ubiquitination, methylation, phosphorylation, and overall acylation), which together influence the gene expression of specific or groups of senescence-associated genes (SAGs)[8−12].
-
For grasses used for forage and turf purposes, both external abiotic (e.g., light, atmosphere, temperature, soil, water, and mowing, etc.) and biotic factors (e.g., animal feeding, tramping, pathogen, and insect attacks) impact the progress of leaf senescence.
Light
-
For plants, light serves as both an energy source and a crucial signal for growth and stress response[13]. Dense planting or shading by neighboring plants can alter the intensity and quality of light, leading to changes in plant growth patterns and hastening leaf senescence[14]. Prolonged exposure to low-light conditions has a negative impact on turfgrass, inducing leaf senescence, and ultimately affecting plant growth and overall turf quality[15].
Atmospheric pollution and elevated CO2 level
-
Tropospheric ozone, also known as ground-level ozone (O3), is a significant air pollutant in the lower boundary layers. Prolonged exposure to high levels of O3 imposes detrimental effects on photosynthesis and carbon allocation, causing premature leaf senescence and ultimately lower grass yield and quality[16,17]. Despite efforts to control anthropogenic emissions, additional sources and factors can also contribute to increasing O3 levels[18]. For example, increased O3 exposure triggers senescence in Briza maxima leaves, resulting in a higher ratio of senescent to green biomass, increased fiber content (particularly lignin concentration), and nutritive value for herbivores[19]. Alfalfa (Medicago sativa) varieties with a slower rate of leaf senescence were found to have greater tolerance to O3[20].
An increase in atmospheric CO2 is the primary cause of global climate change. Over the past century, the rate of CO2 accumulation has steadily increased because of land cover changes and fossil fuel combustion[21]. Elevated CO2 (eCO2) could have positive effects on plants, including an increased C:N ratio, production of secondary metabolites, and higher phytohormone concentrations[22]. eCO2 extends the lifespan of forages during autumn, resulting in an extended growing season and higher biomass yield[23,24]. However, eCO2 also adversely affects the nutritional quality of crops and seeds[22]. Similar changes have been observed in grasses, whereby increased levels of CO2 enhance grass productivity at the cost of decreased nutritional quality of the grasses with higher lignin but lower protein contents[25].
High temperature
-
Greenhouse gas emissions are increasing the average air temperature such that the global mean annual temperatures are predicted to rise 0.3–4.8 °C by the year 2,100 with more frequent and severe heat waves[26]. Elevated temperature benefits the early growth stages of annual crops; however, it also leads to early leaf senescence before the final harvest. For example, the increased temperatures enhance leaf photosynthesis and the above-ground biomass of reed canary grass (Phalaris arundinacea) in the early growing stage but eventually result in early senescence and lower biomass at the end of the growing season[27,28]. Furthermore, severe heat stress cause Chl degradation and severe membrane lipid peroxidation, accelerating leaf senescence[29,30].
Soil water content
-
Limited soil water availability is the main cause of drought stress on plants[31]. Grasses grown under drought conditions show a rapid decline in fresh biomass, leaf water potential, and Chl levels, which is normally coupled with early initiation of senescence[32,33]. Grasses exhibiting prolonged greenness in arid and semi-arid habitats often display high resilience to drought[34].
Extremely high soil water content often caused by flooding is associated with low soil oxygen content that imposes disruptive stress on most land plants[35]. For lowland-grown grasses, the recovery ability from submergence is directly associated with the maintenance of green photosynthetically active leaves. Water-logging-intolerant grasses, such as Dactylis glomerata L. and Bromus catharticus Vahl, exhibit premature leaf senescence and persistent partial stomatal closure during recovery, whereas flood-tolerant species Phalaris aquatica L. maintains leaf greenness and stomatal conductance during recovery[36].
Saline-alkali and toxic metal
-
The worldwide area of saline-alkali land is approximately 0.34 × 109 ha. Soil salinization reduces soil fertility, adversely affects plant productivity, growth, and development and damages agricultural productivity and ecosystem diversity[37]. Premature leaf yellowing is a significant symptom in M. truncatula under salt or alkali stress[38,39].
High toxic heavy metal content in top soils due to mining, exhaustion emissions, sewage irrigation, and other activities is another plant abiotic stress factor[40]. When Miscanthus × giganteus is planted in soil contaminated with Pb, Cu, or Zn, its leaves show decreases in the photosynthetic rate and total photosynthetic capacity, noticeable precocious leaf senescence, and reduction in the plant's growth rate and biomass yield[41].
Fungi and bacteria
-
Plants, along with their associated microbiomes, operate as intricate, multispecies organisms. These associations give them mutual benefits in most cases; however, once pathogenic microbes are dominant, plants often die fast or senesce early[42]. For example, the necrotrophic pathogen Phoma medicaginis var. medicaginis causes alfalfa to develop black spring stems and leaf spots. The symptoms of this infection include black spots on the leaves that later turn yellow[43]. In some cases, certain symbiotic bacteria like Clavicipitaceae (specifically, Claviceps purpurea) form mutualistic relationships with drunken horse grass (Achnatherum inebrians). These microbes can prolong the life cycle of leaves, enhance the activity of enzymes, such as superoxide dismutase, peroxidase, and catalase, and augment proline contents. Additionally, these bacteria reduce malondialdehyde contents while increasing the Chl content and photosynthetic rate in leaves[44]. Additionally, treating alfalfa with exogenous PopW, a harpin protein derived from Ralstonia solanacearum, improves its drought tolerance with altered endogenous hormone levels and expression of genes related to drought stress and leaf senescence[45].
Turf and grassland management
-
Effective management techniques, such as fertilization, irrigation, mowing, and grazing management, can significantly postpone the leaf senescence of forage and turf grasses[46−50]. For example, removing switchgrass panicles can help the grass maintain higher Chl content, photosynthetic efficiency, and activity of key enzymes, such as phosphoenolpyruvate carboxylase and ribulose-1,5-diphosphate carboxylase. This process also increases the content of important plant hormones, like zeatin nucleosides, gibberellins, and indoleacetic acid in flag leaves, thereby delaying leaf senescence[51].
-
In recent years, a number of leaf SAGs were cloned and functionally characterized in forage and turf grass. Here, we summarize SAGs cloned from two leguminous and three grass species in the Poaceae family.
Medicago truncatula
-
Alfalfa is an allogamous, autotetraploid plant with a complex genetic background. M. truncatula is a diploid, self-fertile plant closely related to alfalfa and has been used as the leguminous model plant. According to the comparative transcriptomic data, a total of 545 SAGs were identified from naturally senescent leaves[52,53] and up to 1546 SAGs from salt stress-, alkali stress-, and dark-induced leaf senescence[38,39]. During leaf senescence, Chl degradation is coupled with the remobilization of nutrients in leaves. The deletion of the gene STAYGREEN (SGR, encoding a Chl a catabolic enzyme) in M. truncatula enables the plant to maintain its green leaves and retain a significant amount of Chl during senescence. Furthermore, the induced expression of the MtSGR gene has been observed in leaf senescence caused by various stresses[38,39,53,54].
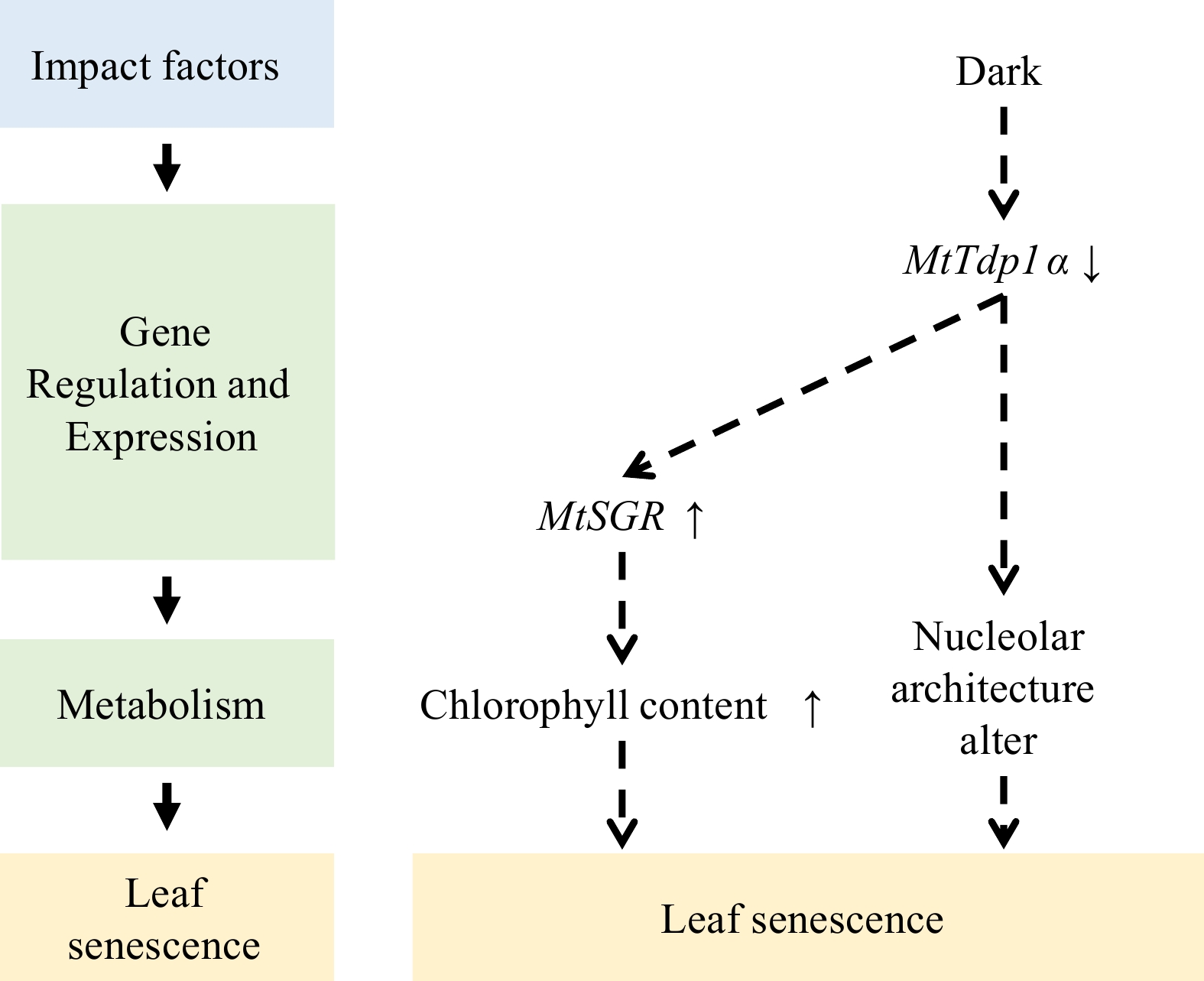
In M. truncatula, mutation of the tyrosyl-DNA phosphodiesterase I (MtTdp1α) gene leads to early leaf senescence. The MtTdp1 protein has two isoforms, MtTdp1α (Medtr7g050860) and MtTdp1β (Medtr8g095490)[55]. Transcriptome analysis reveals that the MtTdp1α downstream genes encompass multiple SAGs, including the MtSGR and SAG101 (an orthologue of Arabidopsis SENESCENCE-RELATED GENE 1 [SRG1]). Silencing the MtTdp1α gene leads to an upregulation of proteolytic genes and genes encoding different subtypes of glutathione S-transferase, as well as the concurrent downregulation of genes involved in photosynthesis. Furthermore, MtTdp1α plays a role in maintaining genomic integrity[56]. In the transcriptome induced by darkness, the expression of MtTdp1α is inhibited[53]. The decreased expression of MtTdp1α caused by darkness weakens the repair function of the genome and promotes the expression of senescence-related genes and leaf senescence (Fig. 1).
Alfalfa (Medicago sativa)
-
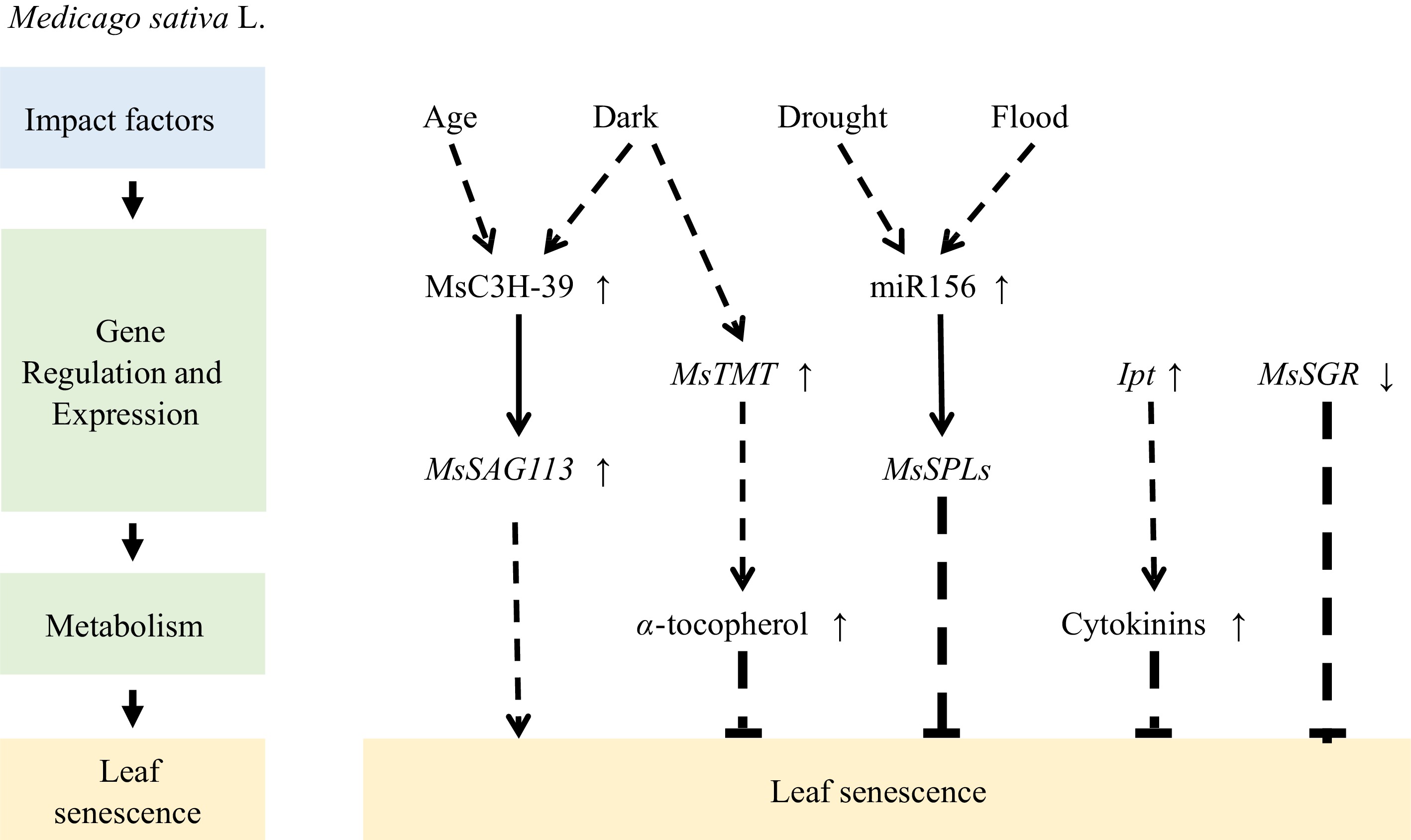
Alfalfa is an important leguminous forage crop cultivated worldwide. The biomass and quality of alfalfa can be affected by leaf senescence. Yuan et al. conducted a transcriptome analysis on mature and senescent leaves and identified 1,250 differentially expressed genes, of which 713 were upregulated, and 537 were downregulated[57]. Calderini et al. constructed a vector containing the SAG12 promoter and a cytokinin biosynthetic gene (IPT) that encodes an isopentenyl transferase. This vector was then used to genetically modify alfalfa and inhibit leaf senescence[58]. The results revealed that senescence occurs at a slower rate in alfalfa SAG12-IPT plants, as indicated by higher Chl levels.
Alfalfa transgenic plants with SGR gene silenced by RNA interference (RNAi) retained over 50% of their Chl content during senescence and higher crude protein content[54]. Another study showed that MsTMT expression is upregulated in the dark. Overexpression of γ-tocopherol methyltransferase (MsTMT) to increase their α-tocopherol contents also delayed leaf senescence and increased crude protein content in transgenic alfalfa[59].
MicroRNA156 plays a crucial role in enhancing alfalfa tolerance to abiotic stress. At least seven MsSPL (SQUAMOSA promoter binding protein-like) genes are regulated by miR156 through transcript cleavage. For example, the miR156/SPL module is involved in drought- and flooding-induced leaf senescence; when miR156 is overexpressed, or its target gene SPL13 is silenced, the onset of leaf senescence caused by flooding is delayed[60]. Similarly, overexpression of miR156 leads to increased drought resistance in alfalfa, whereas silencing its target gene SPL9 delays drought-induced leaf senescence[61]. Thus, the miR156/SPL module may play multiple regulatory roles in stress responses, including stress-induced leaf senescence.
Furthermore, MsSAG113, a negative regulator of the abscisic acid (ABA) signaling, is controlled by various hormones, such as ABA, salicylic acid, and methyl jasmonate (MeJA). Overexpression of MsSAG113 can accelerate the senescence of dark-induced and aged alfalfa leaves, with its downstream genes mostly related to hormone-regulated genes. Therefore, MsSAG113 may play a crucial role in leaf senescence and the hormone regulation network of alfalfa. MsC3H-39 is a CCCH-like zinc-finger protein that directly recognizes and binds to the upstream region of MsSAG113, enhancing its expression. Additionally, the transient expression of MsC3H-39 promotes alfalfa leaf senescence[62], which implies that MsC3H-39 directly regulates MsSAG113 expression and contributes to leaf senescence (Fig. 2).
Perennial ryegrass (Lolium perenne)
-
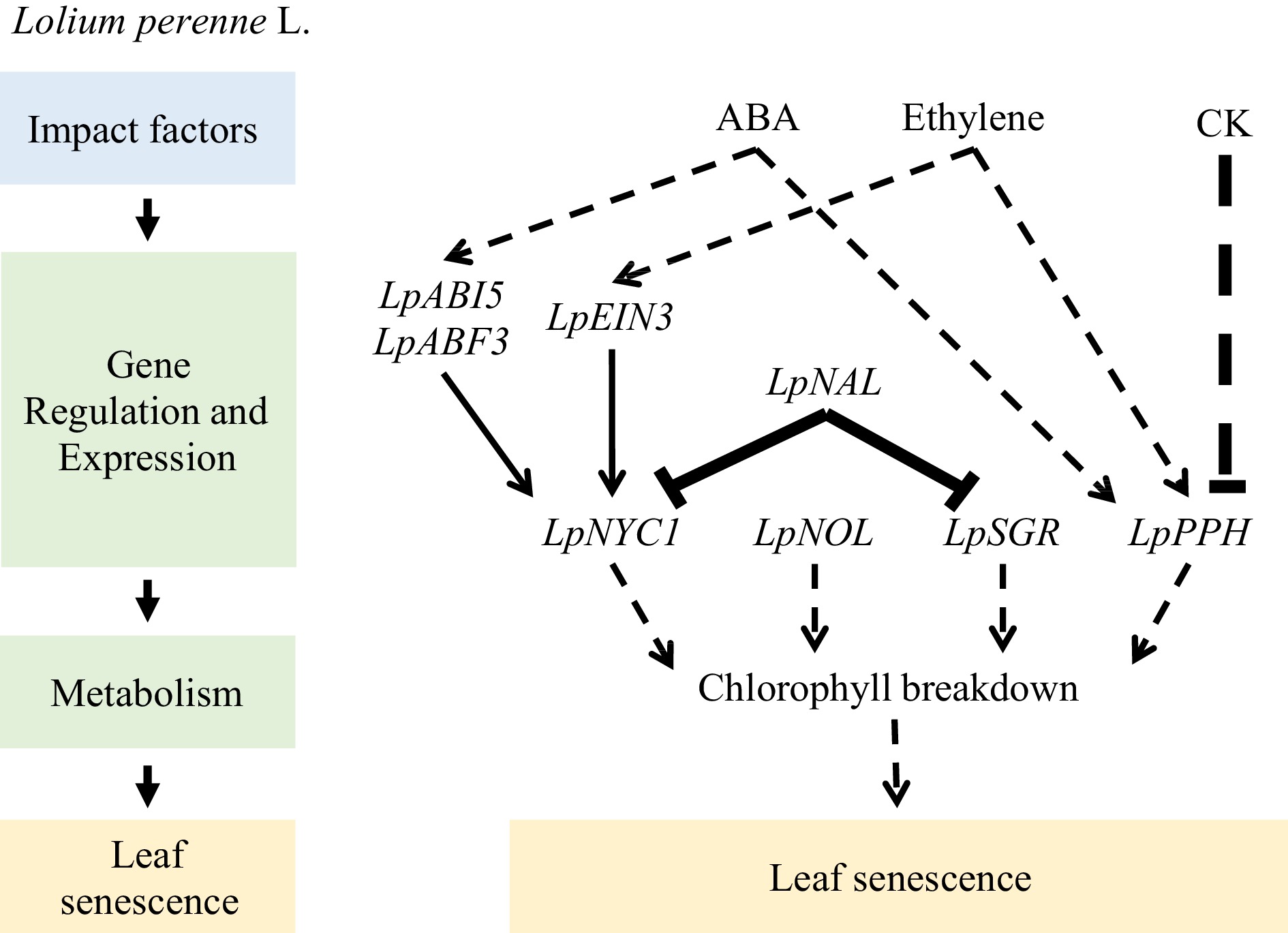
Hormones play crucial roles in regulating leaf senescence in perennial ryegrass. Application of exogenous 6-benzylaminopurine increases the activity of antioxidant enzymes such as superoxide dismutase, catalase, ascorbate peroxidase, monodehydroascorbate reductase, and glutathione reductase and inhibits the accumulation of Na+ in cells, delaying salt-induced leaf senescence[63]. Similarly, exogenous application of melatonin enhances the activity and transcription levels of superoxide dismutase and catalase, activating the superoxide dismutase enzymatic antioxidant pathway and reducing the transcription of genes associated with Chl degradation (LpSGR, LpNYC1, LpNOL, LpPPH, LpPAO, and LpRCCR1) and senescence markers (LpSAG12.1, Lph36, and Lpl69). Up- or down-regulation of Chl catabolic genes, such as LpNYC1, LpNOL, LpSGR, and LpPPH, may accelerate or suppress leaf senescence[64−66]. Moreover, melatonin inhibits dark-induced leaf senescence by downregulating Chl degradation[67]. Additionally, exogenous application of melatonin upregulates the expression of genes involved in cytokinin biosynthesis (LpIPT2 and LpOG1) and their signal response transcription factors (type-B ARRs), downregulates the expression of genes involved in ABA biosynthesis (LpZEP and LpNCED1) and signal transduction (LpABI3 and LpABI5), increases the content of endogenous cytokinins, and reduces the ABA content to alleviate heat stress-induced leaf senescence[68]. The effect of strigolactone on leaf senescence in grass has received little attention; however, exogenous application of a strigolactone analog (GR24) accelerates the downregulation of genes related to Chl biosynthesis (LpHEMA, LpGSA, LpHEMB, and LpCHLH) and promotes the upregulation of genes associated with senescence (LpSAG12.1, Lph36, and Lpl69) and Chl degradation (LpNOL, LpPPH, LpPAO, and LpRCCR1) to promote dark-induced leaf senescence[69](Fig. 3).
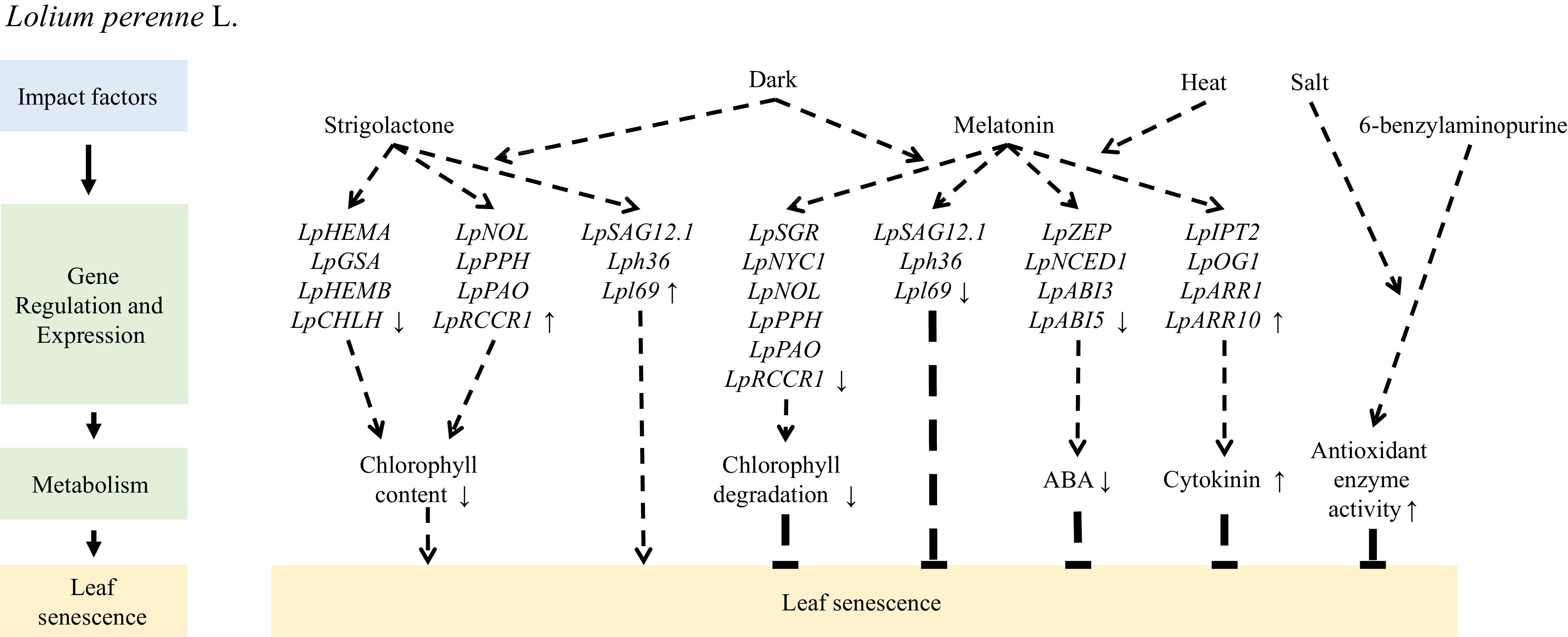
Figure 3.
Mechanism of exogenous hormone application regulating leaf senescence of Lolium perenne L.
In the Chl catabolic pathway, the LpSGR gene, which is responsible for magnesium dechelation of Chl a, is highly expressed in leaves undergoing senescence due to development or darkness. This gene plays a crucial role in Chl degradation, consequently affecting various metabolic processes. In perennial ryegrass, RNAi targeting LpSGR leads to the inhibition of Chl degradation, increasing Chl content and enhancing photochemical efficiency in senescent leaves[70]. However, when subjected to heat stress, the suppression of LpSGR results in the downregulation of genes encoding photosystem proteins and ROS-scavenging enzymes and the upregulation of genes encoding ROS-generating enzymes, thereby accelerating leaf senescence in LpSGR-RNAi lines[71].
Similarly, the Chl b reductases, non-yellow coloring 1 (NYC1)-like (NOL) and NYC1, locate upstream of SGR, whereas pheophytinase (PPH), pheophorbide a oxygenase (PAO), and red Chl catabolite reductase (RCCR) are enzymes downstream of SGR in the Chl catabolic pathway. Previous studies have demonstrated that LpNYC1 is involved in the ABA and ethylene signaling pathways, which can be directly regulated by LpABI5, LpABF3, and LpEIN3[72]. Decreased LpNOL expression or the addition of exogenous Chl promotes Chl accumulation, exhibits an antioxidant effect during heat stress, and delays leaf senescence[73−75]. LpPPH overexpression accelerates Chl degradation. Additionally, LpPPH-mediated Chl breakdown may be positively regulated by ABA and ethylene but negatively regulated by cytokinin[76]. Overexpression of LpNAL leads to a phenotype characterized by delayed leaf senescence or retention of green color, whereas knocking down LpNAL using RNAi accelerates leaf senescence. Furthermore, transcriptome analysis has revealed that LpNAL plays a role in photosynthesis, antioxidant metabolism, ABA and ethylene synthesis and signaling, and Chl catabolism (Fig. 4).
Switchgrass (Panicum virgatum)
-
Switchgrass is a warm-season perennial grass species utilized as both forage for livestock and biofuel feedstock. Improving nitrogen (N) remobilization from aboveground to underground organs during annual shoot senescence is an important goal for the sustainable production of switchgrass as a biofuel crop. Transcriptome and proteome analysis has identified genes that are differentially expressed in various organs during development, such as those involved in protein degradation, nitrogen remobilization, transcriptional processes, and photosynthesis-related proteins[77]. During senescence, the increase in total N content in underground organs is consistent with the decrease in N content in aboveground organs. The contents of 14-3-3-like proteins and glutathione S-transferase proteins in late-senescent leaves are significantly higher than those in early-senescent leaves, indicating that a higher abundance of these proteins might delay leaf senescence[78].
An in-depth understanding of mechanisms controlling mineral uptake, distribution, and remobilization is important for sustainable mineral production. A previous study identified genes associated with mineral transporters in switchgrass and observed relative changes in transporter expression during different growing seasons. Genes from the same family exhibit specific expressions at different stages. Moreover, members of the MRS2/MGT family are responsible for magnesium transport; however, the MRS2/MGT genes expressed during the active shoot growth phase (Pavirv00000269 and Pavirv00025330) differ from those expressed during early senescent (Pavirv00023490 and Pavirv00047094) and fully senescent (Pavirv00069828 and Pavirv00014024) stages[79].
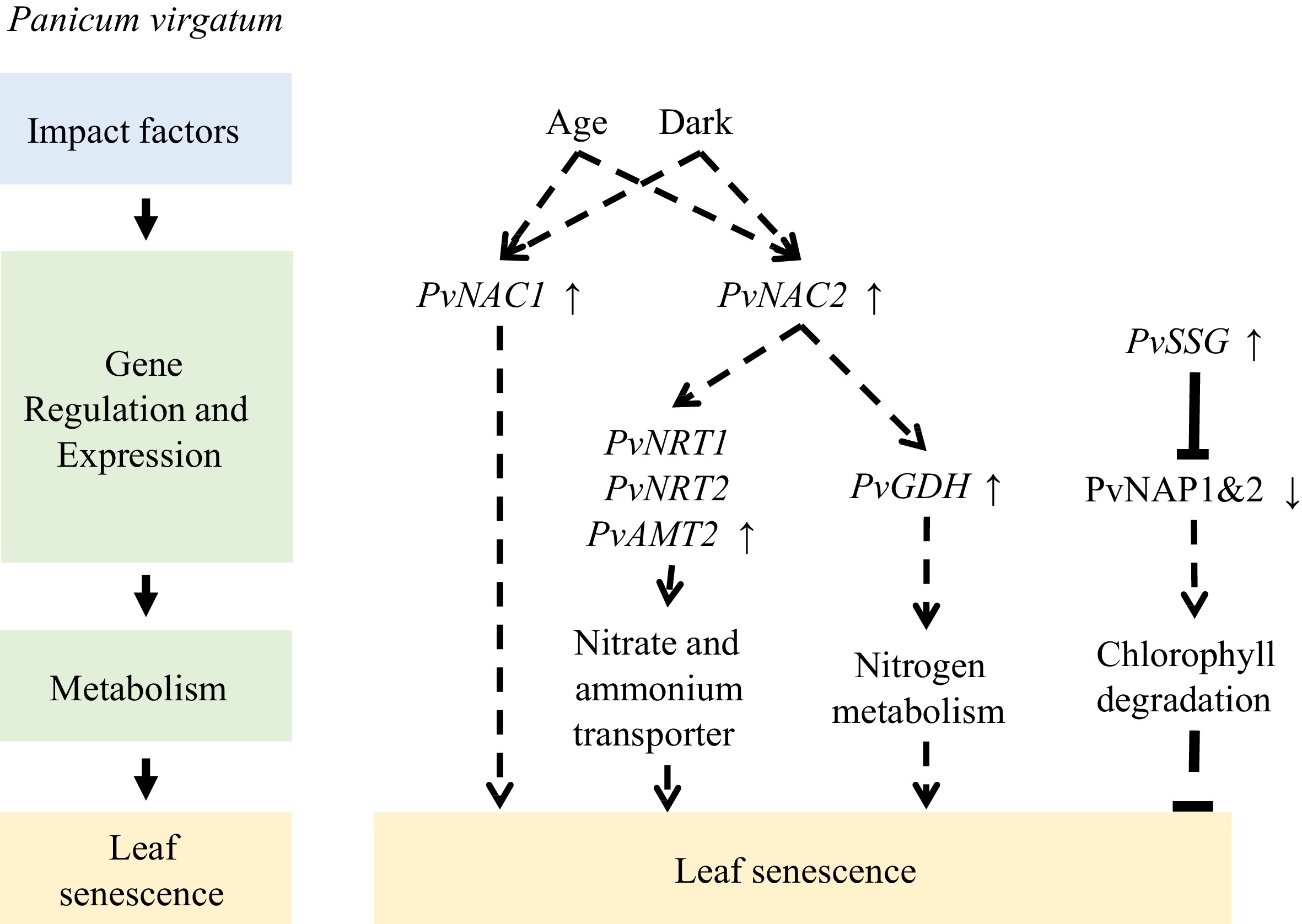
Transcriptome analyses of switchgrass flag leaves revealed the enrichment of transporters during senescence, as well as dynamic changes in other genes and related metabolites during senescence, emphasizing the role of NAC transcription factors in remobilization[80]. In another study, Yang et al. found that the transcription level of PvNAC1 was upregulated by darkness and age[81]. Additionally, PvNAC1 overexpression significantly increased AtSAG12 expression, protein degradation, and nitrogen cycle-related genes, significantly reduced Chl concentration, and promoted Arabidopsis leaf senescence. Conversely, PvNAC2 overexpression in switchgrass resulted in increased aboveground biomass, accompanied by increased transcript levels of the nitrogen metabolism gene PvGDH in leaves and the nitrate and ammonium transporter genes PvNRT1, PvNRT2, and PvAMT2 in roots. In addition to the NAC family, the WRKY family is also involved in switchgrass leaf senescence. According to an analysis of the WRKY family in switchgrass, the expression levels of 23 genes are increased when flag leaves begin to senesce[82].
CCCH-type zinc-finger proteins have been implicated in multiple biological processes and environmental responses in plants. In this context, a switchgrass CCCH-type zinc-finger protein, known as PvSSG, was found to inhibit the binding efficiency of PvNAP1 and PvNAP2 to DNA through protein–protein interactions. Additionally, PvSSG reduces the activation of downstream genes related to Chl degradation by PvNAP1 and PvNAP2 and exhibits inhibitory effects on leaf senescence[83] (Fig. 5).
Creeping bentgrass (Agrostis stolonifera)
-
Creeping bentgrass is a cool-season grass species that actively grows during spring and fall when temperatures range from 15°C to 24°C and declines during hot summer. The decline in turf quality during the summer poses a significant problem for creeping bentgrass in areas with warm climates, particularly in transition zones[30]. Moreover, heat stress reduces Chl content, protease activity, amino acid and total protein content, and antioxidant activity in creeping bentgrass leaves while increasing membrane lipid peroxidation[30,84,85]. Leaf senescence induced by heat stress is negatively correlated with ethylene and ABA accumulation but positively correlated with cytokinin production[86]. The application of exogenous cytokinins decreases the heat-induced membrane lipid peroxidation and protease activity and maintains higher leaf Chl content, Fv/Fm, soluble protein content, and antioxidant enzyme activity. The application of cytokinins also delays leaf senescence and improves turf quality under heat stress[84,87−89]. Several other exogenous reagents have a similar effect on delaying leaf senescence (Table 1).
Table 1. Mechanism of exogenous application of various reagents to inhibit leaf senescence of creeping glumes.
Reagents Treatment Effects Ref. Aminoethoxyvinylglycine Heat Regulating chlorophyll metabolic activities [88, 111] Protease inhibitors in the cysteine, serine, or aspartic classes Suppressing proteolysis [112] Glutamate Suppressing chlorophyll degradation.
Activating amino acid metabolism involved in energy production, antioxidant defense, and nitrogen balance[113] β-sitosterol Alleviating the membrane lipid peroxidation and the enzymatic antioxidant system [114] Chitosan Up-regulating chlorophyll biosynthesis related genes.
Down-regulating chlorophyll degradation and senescence related genes
Increasing the activity of antioxidant enzymes[115] Spermine Maintaining higher chlorophyll content, net photosynthetic rate, photochemical efficiency, and performance index on absorption basis.
Promoting osmotic adjustment ability and antioxidant enzyme activities to enhance the scavenging capacity of reactive oxygen species.
Upregulating transcriptions of heat shock protein genes helping to maintain normal synthesis and functions of proteins[116] Biostimulants (TurfVigor and CPR) Promoting both shoot and root growth [117] Trinexapac-ethyl Improving the chlorophyll content and photosynthetic rate [117] γ-aminobutyric acid, proline, and ammonium nitrate Enhancing chlorophyll content and photochemical efficiency.
Suppressing chlorophyll-degrading enzyme activities under heat stress[118] Nitrogen Enhancing protein abundance in photosynthesis and amino acid metabolism and stress defense systems (heat shock protection and antioxidants) [89] Zeatin riboside Reducing heat-induced membrane lipid peroxidation
Reducing protease activity.
Maintaining higher leaf chlorophyll content, Fv/Fm, soluble protein content and antioxidant enzyme activity[84,87−89] γ-Aminobutyric acid Drought Promoting energy production and conversion, antioxidant defense, and DHN3 accumulation [119] Melatonin Increasing photochemical efficiency, chlorophyll content and relative water content.
Suppressing leaf electrolyte leakage, lipid peroxidation and hydrogen peroxide production.
Up-regulating the cytokinin-signaling and synthesis genes.
Down-regulating the chlorophyll-degradation genes and enzyme activities[120] Cytokinin can effectively inhibit leaf senescence. To obtain transgenic plants with stay green phenotype, researchers transferred the Isopentenyl transferase (IPT) gene, which is responsible for catalyzing a crucial step in de novo cytokinin biosynthesis, into creeping bentgrass. These transgenic plants can delay leaf senescence induced by shade, heat, and drought stress[90,91]. Under stressful conditions, the production of cytokinins in transgenic lines increases, which can help maintain high Chl contents. This positive effect also extends to osmotic regulation, antioxidant enzyme activity, protein abundance, metabolic accumulation, and the preservation of high photosynthesis levels[92−95]. Transcriptomic and proteomic analyses have been used to identify genes encoding proteins related to energy production, metabolism, stress defense, signaling, protein synthesis and transport, and membrane transport, which may play significant roles in the overexpression of IPT in creeping bentgrass[96].
Nitrogen deficiency inhibits plant growth and induces leaf senescence by regulating multiple metabolic processes. Specifically, nitrogen deficiency increases the activity of glutamine synthetase transferase (glutamate dehydrogenase) and decreases the content of amino acids and soluble proteins in leaves[97,98]. However, transgenic lines overexpressing IPT can still inhibit leaf senescence induced by nitrogen or phosphorus deficiency[99]. Therefore, enhancing multiple stress resistance in grasses through exogenous hormones or regulating hormone-related genes may be one of the most efficient methods of delaying senescence.
Furthermore, the membrane proteins involved in ATP metabolism, light harvesting, and photosynthetic photochemical reactions, as well as proteins related to the efficient processing of photorespiratory products and reactive oxygen species, may play important roles in regulating leaf senescence in creeping bentgrass under heat stress[100]. The main reason for the heat-induced decrease in leaf Chl contents is the expression of Chl degradation-related genes and increased chlorophyll degradation enzyme activity[101]. For example, when SGR is silenced in creeping bentgrass, transgenic plants display a green phenotype[102].
Other species
-
Research on leaf senescence has also been conducted on other grass species. Studies have investigated the transcriptome analysis of mature and senescent leaves in red clover (Trifolium pratense L.) and the heat-induced leaf senescence transcriptome of tall fescue (Festuca arundinacea) using RNA-Seq[103,104]. Similarly, transcriptome analysis of dark-induced leaf senescence has been reported in Bermudagrass (Cynodon dactylon L.)[15]. Zoysia japonica has been studied to understand the aging process induced by factors such as age, darkness, and salt, using RNA sequencing[105]. Several genes, including ZjPPH[106], ZjNOL[107,108], ZjSGR[109] and ZjNYC1[110] , have been successfully cloned in Zoysia japonica and overexpressed in Arabidopsis to investigate their roles in senescence.
For convenience, Table 2 presents a comprehensive list of the genes discussed in this paper.
Table 2. List of the gene involved in leaf senescence in grass.
Gene Species Effect Ref. Medicago truncatula Promote [54] Medicago sativa Promote [54] SGR Lolium perenne Promote (except heat stress) [71] Agrostis stolonifera Promote [102] Zoysia japonica Promote [109] IPT Medicago sativa Delay [58] Agrostis stolonifera Delay [90,91] Tdp1α Medicago truncatula Delay [56] TMT Medicago sativa Delay [59] miR156 Medicago sativa Delay [60] SPL9 Medicago sativa Promote [61] SAG113 Medicago sativa Promote [62] NYC1 Lolium perenne Promote [72] Zoysia japonica Promote [110] NOL Lolium perenne Promote [73,74,75] Zoysia japonica Promote [107,108] PPH Lolium perenne Promote [76] Zoysia japonica Promote [106] NAL Lolium perenne Delay [73] NAC1 Panicum virgatum Promote [81] NAC2 Panicum virgatum Promote [81] SSG Panicum virgatum Delay [83] -
Premature leaf senescence reduces the amenity of turf and significantly affects the biomass yield and feedstock quality of forage grasses. The stay-green genotypes have been successfully identified in species like ryegrass and alfalfa and have been integrated into breeding programs to minimize yield losses in plants grown under unfavorable environmental conditions. Compared to model plant species (e.g., Arabidopsis and rice), systematic approaches to unravel the molecular mechanisms underlying leaf senescence are still to be carried out in forage grasses, many questions related remain unanswered.
Outstanding future questions:
(1) What are the specific mechanisms that regulate leaf senescence in forage and turf grasses?
(2) What are the most effective methods for delaying leaf senescence in these grasses?
(3) How can the knowledge gained from research on leaf senescence be applied to molecular breeding?
With the identification of more key genes regulating leaf senescence, combined with the CRISPR gene-editing technology, functional 'stay-green' traits will be achieved in forage or turf grass breeding programs for higher quality, higher yield, and improved stress tolerance.
-
The authors confirm contribution to the paper as follows: study conception and design: Chai M, Zhang K; data analysis: Wen J, Zhang J, Wang ZY, Chai M; draft manuscript preparation: Zhang K, Xie H, Xu B, Chai M. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
This study was supported by the Natural Science Foundation of Shandong Province, China (ZR2023MC120), the National Natural Science Foundation of China (No. 32271755 and No. 31972958) and by National Key Research and Development Program of China (2023YFF1001403).
-
The authors declare that they have no conflict of interest. Bin Xu is the Editorial Board member of Grass Research who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang K, Xie H, Wen J, Zhang J, Wang ZY, et al. 2024. Leaf senescence in forage and turf grass: progress and prospects. Grass Research 4: e004 doi: 10.48130/grares-0024-0002
Leaf senescence in forage and turf grass: progress and prospects
- Received: 17 December 2023
- Revised: 09 February 2024
- Accepted: 13 February 2024
- Published online: 28 February 2024
Abstract: Leaf senescence is a complex biological process regulated by development, phytohormones, and various environmental factors. For forage and turf grasses, controlling leaf senescence can greatly improve forage quality, the amenity of lawn and turf, and the grasses’ stress tolerances. Leaf senescence involves a multitude of gene regulation and metabolic changes, including the alteration of chlorophyll metabolism. Here, we summarized the recent progress of studies on leaf senescence in major forage and turf grass species, such as Medicago truncatula, M. sativa, Lolium perenne, Panicum virgatum, and Agrostis stolonifera, to provide an insight into the development of effective methods for delaying leaf senescence in grass species.
-
Key words:
- Leaf senescence /
- Chlorophyll degradation /
- Stay-green /
- Forage yield /
- Grass and lawns