-
Leaf senescence, the final stage of leaf development, is a tightly regulated process that enables plants to recycle nutrients from aging leaves to support reproductive growth and stress adaptation[1]. This process involves intricate coordination among diverse cell types and transcriptional networks, yet its spatiotemporal dynamics at single-cell resolution remain poorly understood. Traditional single-cell RNA sequencing (scRNA-seq) approaches in plants have been limited by technical challenges, including protoplasting-induced stress artifacts and difficulties in profiling mature or senescing tissues[2−4]. Recent advances in single-nucleus RNA sequencing (snRNA-seq), which bypasses protoplast isolation, have opened new avenues for exploring cell-type-specific transcriptional programs across diverse tissues and developmental stages[5−7].
In this context, Guo et al. present a groundbreaking study titled 'An Arabidopsis single-nucleus atlas decodes leaf senescence and nutrient allocation', published in Cell[8]. Through the optimization of a fluorescence-activated cell sorting-free (FACS-free) single-nucleus isolation workflow, the authors successfully obtained over one million high-quality nuclei, covering 20 tissue types and constructing a single-cell atlas spanning the Arabidopsis life cycle. This atlas resolved 539 transcriptional clusters and 38 cell types, including previously uncharacterized populations such as suberized endodermis cells in roots and trichomes in shoots. By integrating spatial transcriptomics Stereo-seq[9], the study validated novel markers—like pith-specific genes in stems—and revealed conserved cell types (e.g., companion cells, guard cells) alongside organ-specific adaptations, highlighting transcriptional divergence between vegetative and reproductive organs.
Central to the study's innovation is the development of two molecular indices, the senescence-associated genes index (SAG-index) and young-associated genes index (YAG-index), which reflect cellular senescence levels and developmental stages through the weighted expression of core senescence genes and young genes, respectively. These indices unveiled synchronized senescence progression across epidermis, mesophyll, and vascular cells, with spatiotemporal heterogeneity reflecting basipetal senescence patterns (tip-to-base yellowing). Remarkably, scattered 'early-senescing' cells in visually green leaves exhibited stress-response signatures, suggesting microenvironmental triggers of senescence initiation. More importantly, the indices' evolutionary conservation, validated in Populus, underscores their utility beyond Arabidopsis for studying senescence in crops. Moreover, by leveraging weighted gene co-expression network analysis (WGCNA)[10], the authors identified hundreds of potential senescence-regulating hub genes that co-regulated senescence progression. Functional validation using T-DNA mutants confirmed the roles for 50% of these genes in modulating senescence, with perturbations altering chlorophyll retention and marker gene expression.
In addition, the study systematically revealed the spatiotemporal distribution patterns of carbon and nitrogen transporters for the first time. Sugars will eventually be exported transporter (SWEET) sucrose exporters (e.g., SWEET11/12/13) and UmamiT amino acid transporters (e.g., UmamiT18/20/28) exhibited stage- and cell-type-specific dynamics, linking senescence to nutrient mobilization. Sink organs, such as siliques, utilized spatially specialized transporters (e.g., SWEET16/17 in seed coats, UmamiT29 in tapetum) to optimize nutrient partitioning, thereby ensuring the orderly allocation of nutrients from source leaves to sink organs.
This research offers wide-ranging application possibilities, encompassing the two significant areas of basic research and agricultural enhancement. In basic research, the multi-tissue, multi-developmental-stage single-cell atlas and open-source data resources provide a universal framework for analyzing physiological processes such as stress responses and organ formation. For instance, comparing SAG-Index distributions under stress vs normal conditions allows precise identification of stress-induced senescence-related cell subpopulations. In crop improvement, senescence-regulating hub genes, such as WRKY DNA-BINDING PROTEIN (WRKY) family transcription factors, offer new targets for precise control of leaf senescence. Similarly, the spatiotemporal expression patterns of carbon and nitrogen transporters provide specific directions for crop molecular design. The combination of hub genes in the regulation of source-sink relationships and environmental response elements has enabled the rapid creation of environmentally intelligent crop germplasms that can increase yields in favorable conditions and ensure stable yields in adverse conditions[11]. Guo et al.'s study reveals more hub genes in source-sink regulation, providing abundant resources for the design of environmentally intelligent crops.
In conclusion, Guo et al.'s study marks a new phase of systematic integration in plant development research. Through single-nucleus transcriptomics, they not only mapped the multi-organ cell atlas of Arabidopsis but also established methodological paradigms for quantifying senescence dynamics and analyzing nutrient transport networks (Fig. 1). In the future, more combining spatial transcriptomics technologies could localize gene expressions at spatial coordinates at single-cell resolution, elucidating signaling interactions between senescing mesophyll 'hotspots' and the vascular system[9,12]. The introduction of single-cell epigenomics (e.g., ATAC-seq) will reveal the dynamic chromatin accessibility of senescence-related genes, clarifying how WGCNA-identified hub genes regulate downstream pathways through epigenetic modifications[13]. The integration of these technologies will advance plant systems biology research from single-dimensional atlas construction to multidimensional dynamic modeling, ultimately providing precise molecular targets for enhancing crop stress resistance and yield.
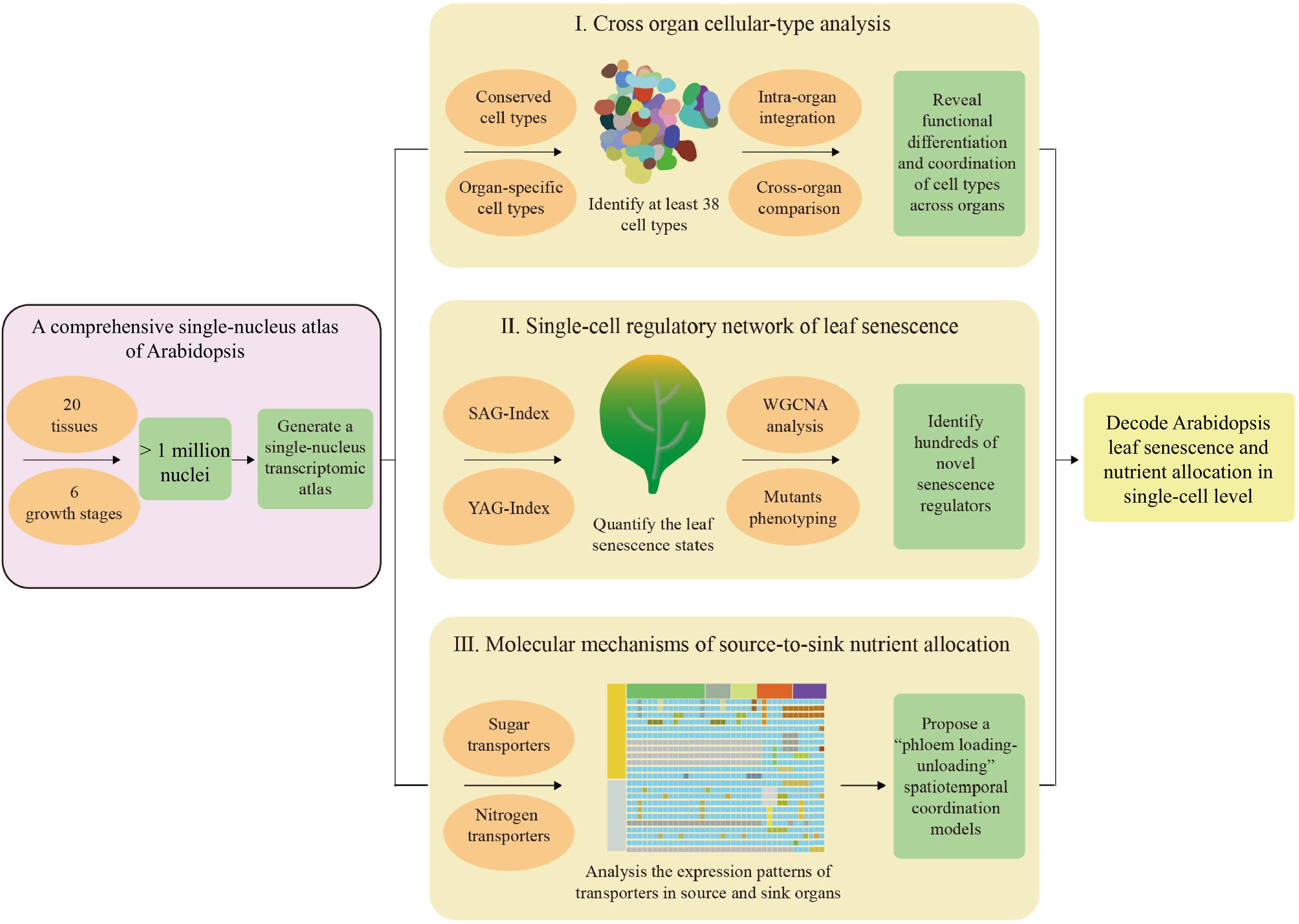
Figure 1.
A new paradigm for studying the dynamic process of leaf senescence at the single-cell level. Firstly, construct a multi-tissue single-nucleus transcriptomic atlas of Arabidopsis. Then, systematically dissect the regulatory mechanisms of leaf senescence from three dimensions: cross organ cellular-type analysis, single-cell regulatory network parsing of leaf senescence, and exploration of the molecular mechanisms underlying source-sink nutrient allocation. Finally, decoding leaf senescence and nutrient allocation.
HTML
This research was supported by the CAS Project for Young Scientists in Basic Research (YSBR-078) and the National Natural Science Foundation of China (32200275).
-
The authors confirm contribution to the paper as follows: manuscript organizing and writing: Yu Y, Xu C. Both authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Yu Y, Xu C. 2025. Decoding the cellular symphony of leaf senescence and nutrient allocation. Seed Biology 4: e007 doi: 10.48130/seedbio-0025-0010 |













