-
Embryos typically originate from the zygote, a totipotent cell produced by the fusion of gametes. Totipotency describes the capacity to generate an entire organism and has long been considered a unique property of the zygote. Nevertheless, plant cells also exhibit remarkable totipotency, which may have evolved as an adaptive strategy to compensate for their immobility and consequent vulnerability to injury. Under suitable conditions, differentiated somatic cells can regain totipotency and begin dividing, giving rise to somatic embryos that are morphologically and physiologically comparable to zygotic embryos[1]. This process is known as somatic embryogenesis. Depending on the species, tissue source, and induction conditions, somatic embryogenesis can occur via multiple cellular pathways. For instance, embryos may arise directly or indirectly (via a callus-mediated process) from a single cell or multiple cells within various tissue layers[2,3]. Since its first documentation in the 1950s[4], somatic embryogenesis has become a powerful tool in plant biotechnology, enabling the propagation of endangered species and the production of transgenic plants with enhanced traits. However, the precise cellular origins of somatic embryogenesis and the molecular mechanisms that enable somatic cells to regain totipotency remains incompletely understood.
In a recent study, Tang et al.[5] demonstrated that somatic embryogenesis induced by LEAFY COTYLEDON2 (LEC2) initiates from specific stomatal progenitors within individual epidermal cells. Their work underscores the essential role of endogenous auxin biosynthesis in driving transcriptional reprogramming and activating embryonic genes during this process. The study identifies a direct developmental pathway from stomatal progenitors to totipotent cells and elucidates how the integration of transcriptional and hormonal signals unlock cellular plasticity. These findings provide new insights into the mechanisms underlying plant regenerative capacity.
In Arabidopsis, somatic embryogenesis can be induced via ectopic expression of LEC2[6]. In this study, Tang et al.[5] employed an estradiol-inducible LEC2-iOX line to induce LEC2 ectopic expression starting from seed germination, which led to the formation of somatic embryos in cotyledon tissues. Using time-course scanning electron microscopy (SEM), the authors observed that these somatic embryos originated directly from individual cotyledon epidermal cells, bypassing an intermediate callus stage. This established a tractable experimental system for investigating the molecular mechanisms underlying direct somatic embryogenesis in vivo. By tracking the co-localization of the embryonic reprogramming marker SOMATIC EMBRYOGENESIS RECEPTOR KINASE1 (SERK1) with specific stomatal lineage markers, the researchers identified that SPEECHLESS (SPCH)-expressing meristemoid mother cells (MMCs) within the stomatal lineage serves as the source of totipotent somatic embryo founder cells (SEFCs)[7−9]. They further demonstrated that both auxin biosynthesis and SPCH expression are essential for the transition from MMCs to SEFCs following LEC2 induction. Expression of SERK1 in SEFCs becomes detectable approximately 96 h post-induction. Prior to this, localized auxin biosynthesis—visualized using YUCCA4::GFP (YUC4::GFP) and DR5::GFP reporters—commenced at approximately 84 h. Inhibition of auxin biosynthesis completely abolished somatic embryo formation, as did mutation of the SPCH gene. Based on these findings, the researchers further investigated whether LEC2 and SPCH coregulate auxin biosynthesis. They discovered that LEC2 interacts with SPCH to synergistically cis-regulate the expression of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and YUC4, significantly enhancing auxin biosynthesis. This mechanism facilitates localized auxin accumulation, thereby promoting the fate transition from MMCs to SEFCs.
To systemically analyze the cell-type-specific transcriptional dynamics and lineage plasticity during direct somatic embryogenesis, the authors performed snRNA-seq on cotyledons collected at approximately 60, 72, 84, 96, and 108 h post-induction. The cells were initially classified into 12 transcriptionally distinct clusters, with clusters 8–10 corresponding to stomatal lineage-like cells and cluster 7 exhibiting a somatic embryo-like identity. Using laser capture microdissection combined with RNA sequencing (LCM-RNA-seq), the researchers generated transcriptional profiles of isolated SEFCs[10], which confirmed that cluster 7 most closely matched the SEFC transcriptome signature. Subsequently, pseudotime analysis was conducted by integrating clusters 8–10 with cluster 7, constructing a developmental trajectory from the MMC/M-like state through a guard mother cell (GMC)-like intermediate state toward totipotent SEFCs. This approach provides a clearer delineation of the bifurcation point between stomatal differentiation and somatic embryogenesis. Furthermore, through transcription factor network analysis, 11 potential core transcription factors regulating this switch were identified. Among these, LEC2, SPCH, MP, and LBD16 were functionally validated in this study. These findings establish a foundational framework for further investigating the multi-layered regulatory networks that initiate reprogramming during this process.
The precise cellular origins of somatic embryos have long remained poorly understood. This study establishes a tractable experimental system where somatic embryos develop directly from individual cells, allowing for a detailed investigation into their origins. Through this approach, the researchers discovered that totipotent SEFCs originate from SPCH-expressing MMCs within the stomatal lineage following LEC2 induction in Arabidopsis cotyledons. By employing live imaging of fluorescent reporter lines that specifically label distinct developmental stages of the stomatal lineage, the researchers precisely identify the temporal window during which the fate bifurcation point emerges after LEC2 induction, providing direct visual evidence of cellular identity transition. Furthermore, through integrated analysis of snRNA-seq and LCM-RNA-seq data, the study clearly delineates the identities of distinct cell clusters and reconstructs the developmental trajectory from MMCs to SEFCs. Comparative transcriptomic analysis of cell clusters near the fate bifurcation point offers a comprehensive perspective on gene expression dynamics during cell fate transition. This study establishes an exemplary framework for deciphering the cellular origins of somatic embryogenesis, delivers mechanistic insight into how developmental and hormonal signals coordinate to unlock totipotency, and provides a fresh perspective on how a single somatic cell regenerates an entire plant[11] (Fig. 1). Furthermore, the mechanisms uncovered here—such as modulating SPCH expression—hold potential for improving the efficiency of somatic embryo regeneration, which could accelerate applications in endangered plant conservation, genetic engineering breeding, and synthetic embryogenesis.
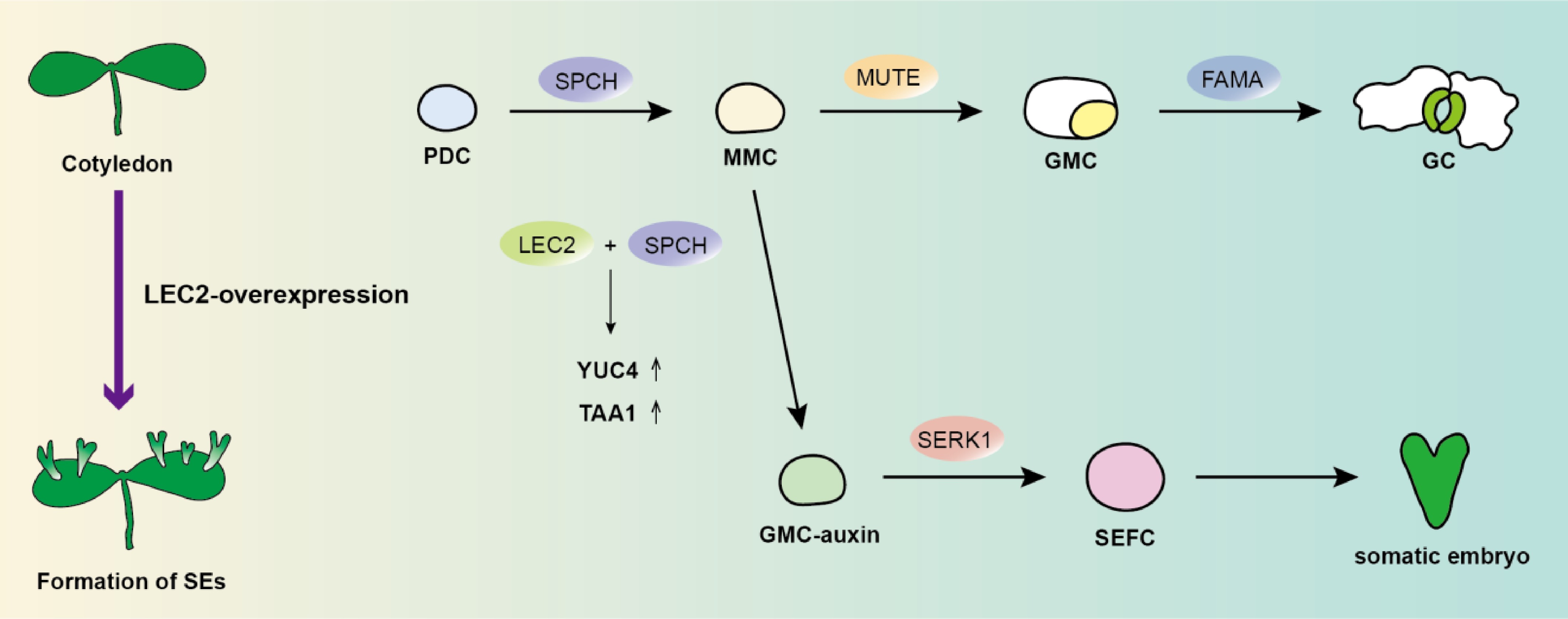
Figure 1.
Trajectory model of cellular reprogramming toward totipotency during LEC2-mediated single-cell-initiated somatic embryogenesis. In Arabidopsis normal stomatal development, SPCH-expressing meristemoid mother cells (MMCs) differentiate from protodermal cells (PDCs) and subsequently progress through the guard mother cells (GMCs) stage to form mature guard cells (GCs). Upon LEC2 overexpression, LEC2 and SPCH cooperatively activate TAA1 and YUC4. This cascade induces transcriptional reprogramming in SPCH-expressing MMCs, diverting them from the default stomatal lineage fate and redirecting their identity toward totipotent somatic embryo founder cells (SEFCs).
Nevertheless, several important questions remain open for future investigation. First, the applicability of the mechanism elucidated in this study to other somatic embryogenesis systems requires further validation. For instance, while somatic embryogenesis in Arabidopsis can be induced through ectopic expression of various transcription factors, such as WUSCHEL[12], AGAMOUS-LIKE15[13], and BABY BOOM[14]. It remains unclear whether these pathways employ similar cellular reprogramming logic to initiate cell fate transitions. Moreover, the generalizability of the proposed mechanism across diverse tissue types, species, and environmental conditions warrants systematic exploration. Second, although the study identified several promising core regulators through transcription factor network analysis, the detailed molecular circuitry through which these factors orchestrate somatic embryogenesis—including their downstream targets, epigenetic interactions, and post-translational modifications—has yet to be fully elucidated.
HTML
The authors' lab is supported by the National Key R&D Program of China (Grant No. 2023YFE0101100), and by the National Natural Science Foundation of China (Grant No. 32230010).
-
The authors confirm their contributions to the paper as follows: manuscript organizing and writing: Cao Z, Jiao Y. Both authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Cao Z, Jiao Y. 2025. From stomatal lineage to totipotent embryo: a direct path revealed by LEC2-induced reprogramming. Seed Biology 4: e016 doi: 10.48130/seedbio-0025-0019 |













