-
Major chronic diseases such as cancer, cardiovascular, cerebrovascular, neurological, and metabolic diseases have become the main diseases threatening the health of humans in modern society[1−4]. Despite the success of new drugs entering into the market, it is widely acknowledged that many diseases lack satisfactory therapeutic drugs. Eroom's law, which was proposed in 2012, to describe the high cost and low productivity of the drug R&D pipeline despite technical advances[5], has prompted reflections on the dogma of drug discovery. Among the potential contributing factors underlying the dilemma, the authors highlighted a progressively higher bar for improvements over existing therapies, and a progressive lowering of risk tolerance by regulatory agencies. It has also prompted the collaboration between traditional pharmaceutical companies and artificial intelligence (AI) firms to address the challenges. However, to date, there has been a low success rate in the stage of clinical trials due to insufficient clinical benefits.
What are the key reasons for this daunting picture, and how do we solve the unmet clinical needs? From a biological viewpoint, it is acknowledged that the etiology and pathological mechanisms of chronic diseases are complex, involving interactions between multiple links, cells, and organs[6]. For example, the redundancy and compensation pathway in proliferation allows tumor cells to survive and recur despite drug intervention, and the robust network of metaflammation and cell death in metabolic dysfunction-associated steatohepatitis (MASH) partially explains why the inhibition of a single target may fail to confer sufficient clinical benefits. These biological underpinnings, as further discussed below, contribute to a dilemma of unsatisfactory efficacy in clinical trials and a high attrition rate of new drug development. At the same time, the existing drug development pipeline is highly concentrated on around 1,000 classic targets such as G protein-coupled receptors (GPCRs), kinases, ion channels, etc., which showed advantages in terms of historical experience in druggability, well-defined chemical characteristics, and extensive clinical validation. However, in consideration of 3,000−10,000 estimated potential druggable targets, this overlapping also depicts a picture of target exhaustion[7], with many novel targets and innovative drugs remaining to be elucidated. Moreover, the 'one gene, one target' philosophy contributes to the concept of 'undruggability', which may stem either from the inability to modulate a target protein using traditional approaches, or from the insufficiency of regulating a single node alone in complex networks to produce favorable therapeutic effects. These difficulties are intertwined, collectively challenging the classical paradigm of single target-specific drugs. Taken together, the shortage of druggable targets and modulatory strategies has become an important bottleneck restricting global new drug development, suggesting the need to pursue multi-target therapeutic approaches for chronic diseases.
-
Chronic diseases largely implicate multiplexed mechanisms, and targeting only one node of signal pathways is insufficient to combat the complex phenotypes. For example, tumor cells can inhibit T-cell activity through the PD-1/PD-L1 pathway and, at the same time, activate the inflammatory microenvironment (such as secreting IL-12) through the NF-κB pathway, thereby promoting immune evasion[8]. The signal integration of these two pathways is a key mechanism of immune suppression. Indeed, the treatment of chronic diseases by a combination of multiple drugs has become increasingly popular in the clinic. In recent years, the FDA has shown an upward trend in approving fixed-dose combination (FDC) drugs (e.g., Entresto, Cobenfy) (Table 1). Clinical trials have also shown that statins, ezetimibe, and fibrates can be safely combined to augment the lipid-lowering efficacy[9]. Also, rapid advances in the field of cancer biology have facilitated the development of novel combination therapeutics targeting metabolic, epigenetic, and immune mechanisms that are hijacked by cancer cells to support tumor growth and progression[10,11]. Accumulating clinical trials have been launched to test epigenetic agents in combination with chemotherapies, immunotherapies, or other targeted agents for improved treatment response (Table 2). Most importantly, the 2025 Lasker Award recognizes the cystic fibrosis (CF) triple-drug therapy, which turns this fatal disease into a manageable one and saves patients' lives.
Table 1. Summary of clinically approved combination drugs.
Trade name Time to market
(FDA approval)Ingredient Indication Targets Widaplik 2025 Telmisartan, Tmlodipine, Indapamide Hypertension Angiotensin II receptor, L-type calcium channel, NCC Cobenfy 2024 Xanomeline, Trospium chloride Schizophrenia M1, M4 Inavolisib 2024 Palbociclib, Fulvestrant Breast cancer PI3Kα, hormone receptor, HER2 Opdualag 2022 Nivolumab, Relatlimab Metastatic melanoma PD-1, LAG-3 Entresto 2015 Sacubitril, Valsartan Heart failure Neprilysin, AT1 Evotaz 2015 Atazanavir, Cobicistat HIV-1 HIV protease Namzaric 2014 Memantine hydrochloride,
Donepezil hydrochlorideAlzheimer's disease AChE, NMDA receptor Dapagliflozin/metformin 2014 Dapagliflozin, Metformin hydrochloride Type 2 diabetes mellitus SGLT2, AMPK Symbicort 2006 Budesonide, Formoterol fumarate COPD, Asthma GR, β2-AR Table 2. Representative example of combination drugs under clinical trial (2023–2025).
Name Time to market
(FDA approval)Launch year Indication Targets Ref. Letrozole 1997 2025 Alzheimer's disease APOE, SYT1, TREM2, GFAP [75] Irinotecan 1996 Prazosin 1976 2023 Traumatic
brain injuryα1, α2, β1, β2 [54] Atipamezole 1966 Propranolol 1967 Rapamycin 1999 2024 Neuroblastoma mTOR,
BCR-ABL, Topoisomerase I[28] Dasatinib 2006 Irinotecan 1996 Temozolomide 1999 Navitoclax / 2024 lymphoblastic leukemia MDM2, Bcl-2, Bcl-xL, Bcl-w [9] Idasanutlin / Durvalumab 2017 2024 NSCLC CTLA4, PD-L1 [130] Tremelimumab 2022 Azacitidine 2004 2024 Peripheral T
cell lymphomas (PTCLs)DNMT, HDAC [33] Chidamide / Ipilimumab 2011 2025 Non-clear
cell renal cell cancers (nccRCCs)CTLA-4, PD-1 [10] Nivolumab 2014 Ibrutinib 2013 2025 CNS lymphoma PD-1, BTK [23] Nivolumab 2014 The concept of polypharmacology, first used by Domino in 1971 to describe where a single compound has activating or inhibitory effects on multiple target proteins[12], also supports the promise of multi-target regulation in clinical therapy. Currently, there are 10 approved multi-target drugs on the market, including seven anti-tumor drugs, and one antidepressant drug[13]. As a typical case, ample clinical data have shown that statins, used to treat hypercholesterolemia, and metformin, the first-line medication for type 2 diabetes, rely on simultaneous modulation of multiple targets to confer beneficial metabolic effects[14]. Also, the tricyclic antidepressants are believed to be 'dirty' drugs, since they not only block the neuronal serotonin and noradrenaline transporter, but also act on a series of GPCRs, including muscarinic, histamine H1, and α1-adrenergic receptors[15]. As a dual agonist of GIPR and GLP-1R, tirzepatide can simultaneously target two synergistic nodes in metabolic regulation, producing superior efficacy compared to agonists that act solely on GLP-1R (such as semaglutide)[16]. Moreover, several studies show that two drugs covalently modifying different residues of the same protein can produce deeper conformational collapse and superior functional knockout (intra-target synergy). This concept is especially relevant to natural products (e.g., Rhodiola crenulata, Pu-erh tea, Sappanwood) that furnish multiple electrophilic fragments directed against viral proteases or host signaling hubs[17−19]. Collectively, these examples underscore the therapeutic potential of both inter- and intra-target polypharmacology, thereby highlighting the broader utility of natural products as promising sources for the development of next-generation multi-target drugs.
The intrinsic adaptive rules of the cellular signal network
-
Mammals have evolved sophisticated mechanisms to maintain homeostasis, adapt to environmental changes, and ensure proper physiological functioning. At the cellular level, these rules largely encompass substitution, compensation, redundancy, and feedback, all of which play a unique role in sustaining homeostasis in the face of environmental perturbations. Nevertheless, these characteristics of the biologic signaling network may also contribute to the insensitivity of drug treatment[20,21]. Highly specific interference of a certain signal node is likely to trigger adaptive change of biological signals, highlighting the need for multi-target therapeutics[22]. Although these biological rules also apply to the treatment philosophy of acute conditions (e.g., antimicrobial combination, anesthetic adjuvants), they are particularly advantageous for chronic disease conditions due to their prolonged timeline, which allows compensatory network responses to fully manifest, and the temporal interconnectedness of their pathogenic networks, as discussed below.
Substitution refers to the ability of one biological signal or pathway to take over the function of another when the latter is impaired or unavailable[23]. Compensation involves adjustments in related biological signals or systems to counteract changes or imbalances[24,25]. Essential for cellular adaptation, these mechanisms could be co-opted by tumors for adaptation and survival, as illustrated by a previous study showing the synergistic effect of LAG-3 and PD-1 in promoting exhausted CD8+ T cells in infections and tumors[26].
Redundancy is a crucial adaptive strategy where multiple biological signals or pathways perform similar functions[27]. This duplication mechanism, however, has also been hijacked by tumors in response to chemotherapeutics[28]. For example, focal adhesion kinase (FAK), a non-receptor tyrosine kinase, exerts a pivotal role in the drug resistance of tumor cells[29]. The signaling network of FAK exhibits significant redundancy, since it can interact with integrins, growth factor receptors (e.g., EGFR, PDGFR), cytokine receptors, and other molecules, thereby activating multiple downstream pathways. When FAK is subjected to single-agent inhibition, cancer cells can achieve compensation via alternative pathways (Fig. 1).
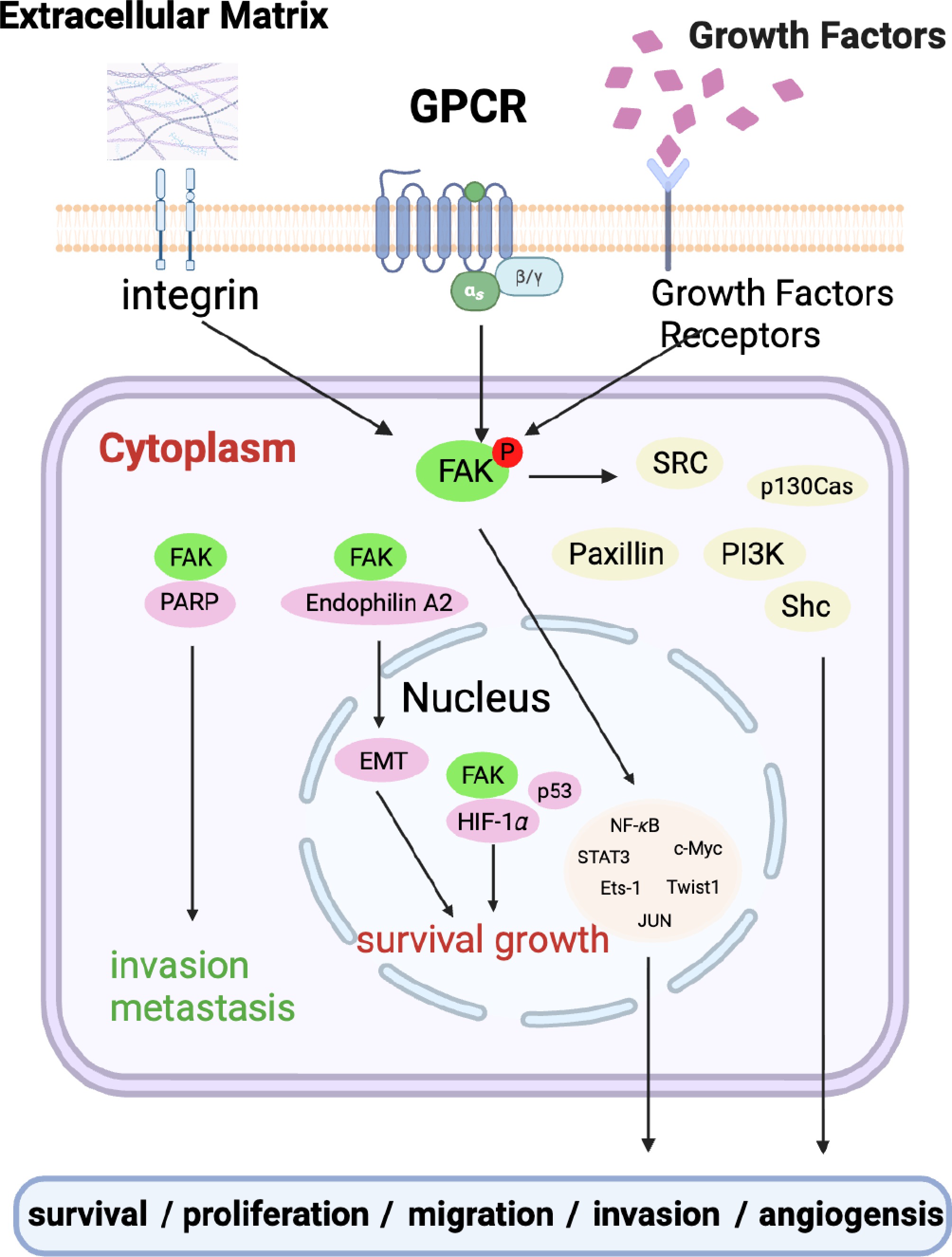
Figure 1.
Schematic showing the redundant signaling pathway employed by tumor cells for survival. In the diagram, extracellular matrix, GPCR, growth factors, etc. activate FAK, triggering a series of intrinsic adaptive regulatory mechanisms such as intracellular signal transduction, which in turn regulate processes like cell survival, proliferation, and migration.
Feedback mechanisms are integral to the balance of biological signals, which involves the interpretation and adjustment of output signals based on their input. This also explains the increasing popularity of combinatory therapy for tumors. For example, anti-vascular endothelial growth factor (anti-VEGF) agents, including bevacizumab, have been shown to induce hypoxia in tumor cells after the inhibition of tumor angiogenesis. This hypoxic microenvironment, in turn, triggers the activation of the epidermal growth factor receptor (EGFR) signaling pathway. Specifically, hypoxia-mediated upregulation of EGFR expression occurs via hypoxia-inducible factor-1α (HIF-1α), which enhances the invasive and metastatic potentials of tumor cells[30]. Consequently, this molecular cascade counteracts the therapeutic efficacy of anti-angiogenic interventions, which warrants dual-agent chemotherapy to overcome insensitivity.
Multi-layer signal integration and diversification
-
Cells and tissues perceive a multitude of signals from their micro- and macro-environment, and they must integrate these inputs to make appropriate decisions. This integration can occur at various levels, from the molecular interactions within a cell to the communication between different organs[31−33]. Homeostatic signaling networks represent the intricate molecular and cellular systems that maintain biological stability across diverse physiological contexts, enabling organisms to buffer fluctuations in internal and external environments. These networks integrate multiple layers of regulation, collectively ensuring the robustness of biological systems[34,35]. This may also explain why high-specificity/affinity drug design is of high risk for unwanted on-target side effects.
The promiscuity of ligand-receptor interaction and downstream propagation is a cornerstone of biologic network that allows for a fine-tuning of cellular responses[36,37]. The aryl hydrocarbon receptor (AhR), for example, plays an important role in maintaining cellular homeostasis and pathophysiology, and there is increasing evidence that the AhR interacts with a myriad of endogenous and exogenous modulators (agonists and antagonists) at differential affinity that exhibit tissue/cell-specific functional diversity[38,39]. From an evolutionary perspective, the promiscuity allows a single protein to integrate multiple environmental cues, amplifying the network's capacity to adapt to changing conditions while maintaining specificity through ligand affinity and cellular localization[40].
The gut microbiota constitutes a critical hub of signaling networks, particularly through its perception of diverse environmental cues to influence metabolic, neural, and immune pathways[41]. Commensal bacteria ferment indigestible substrates to produce short-chain fatty acids (SCFAs), bile acids, and neurotransmitters, which act as signaling molecules to regulate local and distal organs[42], as best exemplified by the bidirectional interaction via the gut-brain axis (Fig. 2). In this fashion, metabolites from the host-microbe metabolic network have emerged as signaling molecules orchestrating the responses across cells and tissues. Key signaling molecules such as bile acids have been widely shown to coordinate the information fluxes between tissue-resident cells and immune cells, which is attributed to their interaction of diverse receptors/sensors on different cells[43]. The signaling network behind bile acids and other host-microbe co-metabolites has been regarded as a potential target for the treatment of chronic diseases, as reviewed elsewhere[44−46]. On the other hand, herbal medicines, which is a combination of multiple phytochemicals, are widely believed to treat chronic diseases by gut microbiota modulation, especially given their extensive exposure to these commensals in the gut[47,48]. The mechanism largely involves restoring the ecological balance of gut microbiota (e.g., enriching SCFA producers such as Faecalibacterium prausnitzii) and improving microbiota-mediated metabolic and immune regulation[47], which provides a new idea of 'host-microbe' dual regulation for chronic diseases.
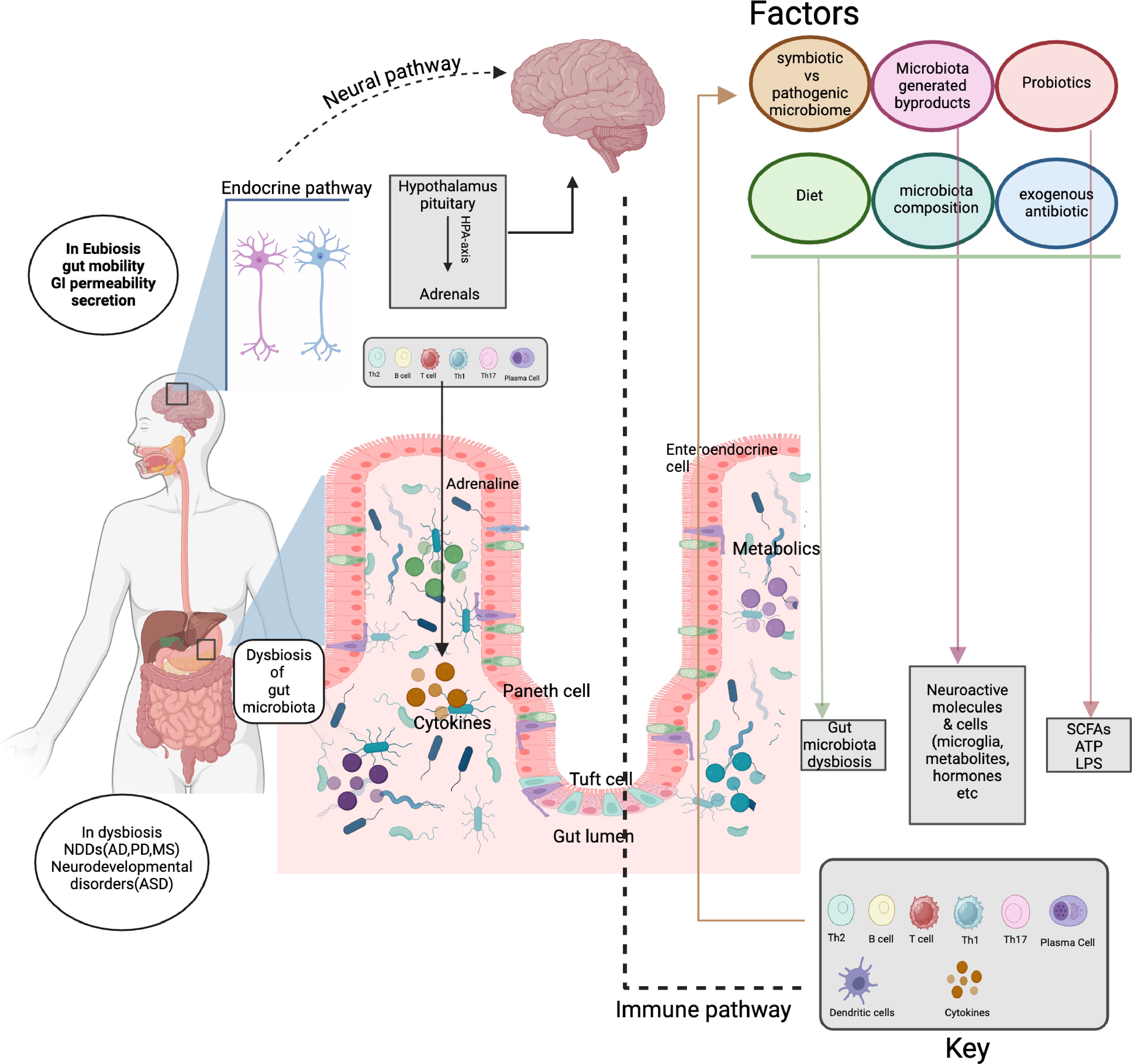
Figure 2.
Schematic showing gut microbiota as a signaling hub in the bidirectional crosstalk between the gut and brain. Gut microbiota: Interacts with factors (diet, probiotics, antibiotics) to impact composition, byproducts, metabolism, and immunity. Immune system: uses immune cells (T cells, B cells, and plasma cells) and cytokines to mediate gut-body communication. Endocrine and nervous systems: enable gut-brain communication via the hypothalamus-pituitary-adrenal (HPA) axis and neural pathways, influencing gut and neural functions.
Trans-organ communication and comorbidity
-
Multi-organ comorbidity, which refers to interconnected dysregulation of multiple biologic signals spanning several tissues and organs, is a typical feature of many chronic diseases. In the context of multi-organ comorbidity, the intricate network of biological signals between different organs becomes disrupted, leading to a cascade of physiological imbalances[49−51]. For example, in patients with diabetes and cardiovascular disease, abnormal insulin signaling not only affects glucose metabolism in the liver and muscles but also impairs vascular function in the heart and blood vessels[52,53]. However, the combined use of metformin, GLP-1RA (such as semaglutide), and statins is an excellent treatment modality to confer clinical benefit. The cardioprotective effect of GLP-1RA complements the lipid-lowering and anti-inflammatory effects of statins, while metformin decreases the potential risk that insulin may exacerbate atherosclerosis.
Moreover, in the progression of chronic diseases, the spatiotemporal heterogeneity of signals across multiple organs becomes dysregulated, and the integration of signals fails to coordinate physiological functions effectively, further contributing to the progression of comorbidities[54,55]. As a metabolic hub, the liver displays aberrant spatiotemporal regulation of glucose-lipid metabolism and neuroimmune signals during the progression of MASH—this not only induces local pathologies like hepatocyte death and astrocyte activation, but also disrupts intestinal barrier integrity and cardiac energy supply through liver-intestine and liver-heart axes, perpetuating a vicious cycle of multi-organ dysfunction[56,57]. This raises questions about the extent to which approved current anti-MASH drugs (e.g., Resmetirom) could confer systems impacts to combat multi-organ comorbidity. Understanding the interplay between biologic adaptation rules and multi-organ comorbidity is crucial for developing novel therapeutic strategies to target multiple disease pathways simultaneously.
-
Homeostatic signaling networks integrate multi-layer, dynamic signaling to maintain biological equilibrium. Such complexity ensures robustness against internal and external challenges, but also underlies the lack of efficient drugs for many chronic diseases[58]. Understanding these interconnected mechanisms is essential for developing next-generation therapies to reinstate homeostasis from the complex pathological states. Given the multi-node characteristics of complex diseases, single drugs are often insufficient to fully halt disease progression[59]. Thus, there is an unmet need to develop 'combined intervention modalities' that act synergistically on several interconnected targets through multiple mechanisms[60].
Here, in this inaugural issue of Targetome, we propose a concept of 'targetome', which entails the complete collection of molecular targets (e.g., proteins, RNA, or DNA) that could be therapeutically modulated to produce synergistic prevention/treatment effects on a specific disease. Targetome could either be the combination of two or more different targets at the same or different cells/organs, or a combination of regulatory nodes on the same target. In light of the essence of this definition, a disease-centered definition of targetome could be the key functional signal nodes playing essential roles in the pathological development and progression of certain diseases and can be manipulated simultaneously to fulfill synergistic and better clinical benefits. In parallel, a drug-centered definition could be the functional signal nodes that are engaged by certain effective therapeutic drugs to elicit clinical benefits.
Combination drugs, by definition, are a combination of drug entities with synergistic effects, such as enhancing efficacy or reducing toxicity for specific diseases. It could be developed as a single drug molecule (pharmacophore combination, bispecific antibody, etc.) targeting two targets or the combination of two or more drug molecules. In relation to the targetome, as is proposed here, the superiority of combinatory drugs lies in the modulation of biological targets that are disease-related and have the potential for synergistic intervention. Polypharmacology, which focuses on the multiple-target mechanism of an exerting therapeutic such as aspirin and sorafenib, could be a protypical example of such a definition. From the molecular level, the concept of 'targetome' serves as the fundamental objective for polypharmacology, but more broadly incorporates system-level dissection of the biologic networks to guide multi-node drug discovery. The core logics and features of these two concepts within the systems-level framework of disease pathogenesis and target discovery are depicted in Fig. 3.
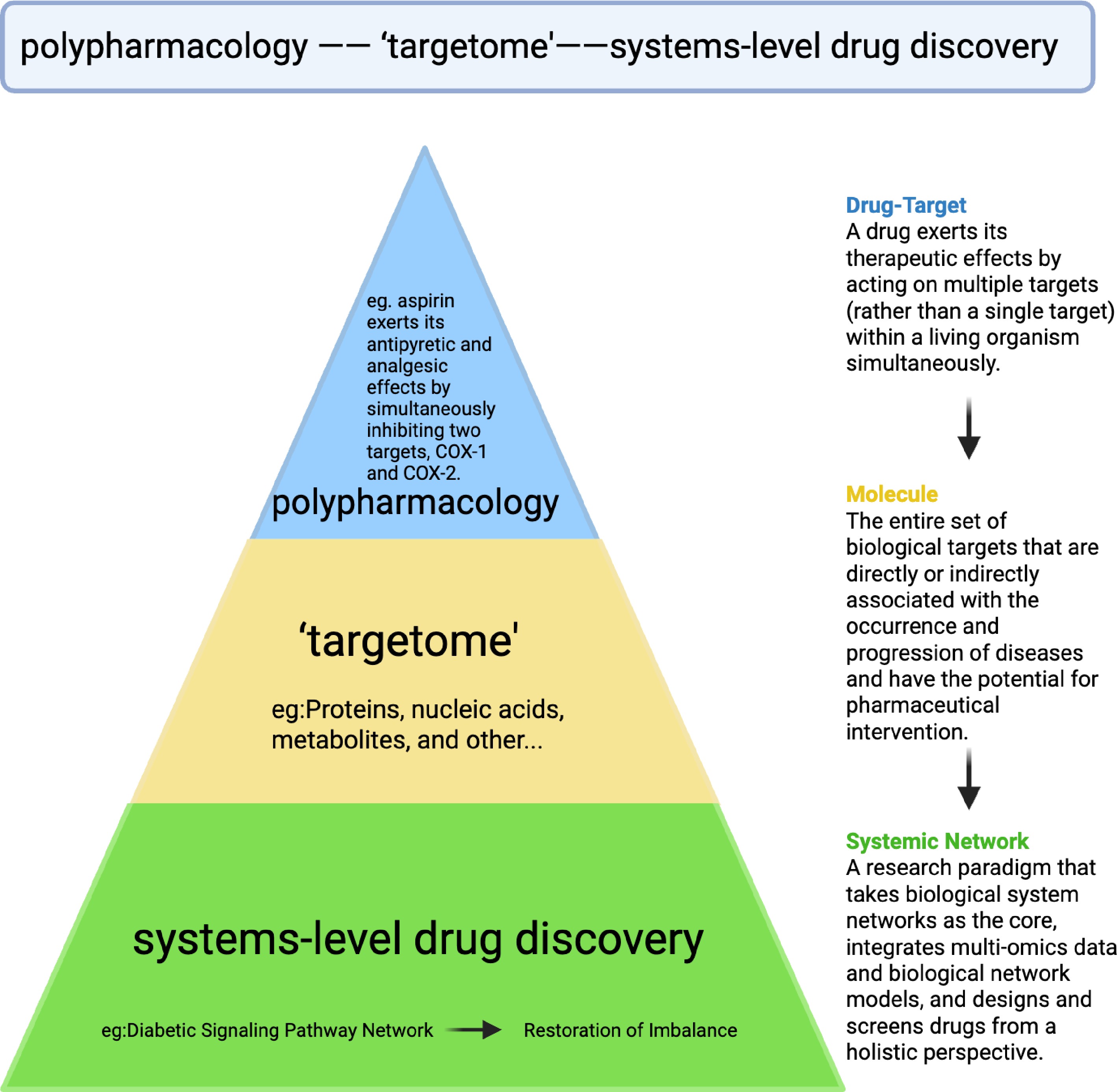
Figure 3.
Schematic illucidation of the relationship between targetome and polypharmacology. Within the triangular systems-level drug discovery framework, 'targetome' represents a much broader concept in comparison with polypharmacology (multitarget drug action, e.g., aspirin on COX-1/2), both of which fit into the system-level strategies in dissecting the disease pathway networks, and holistic drug development.
Traditional drug combinations could be found in several forms. For example, multi-functional antibodies such as Amivantamab are genetically engineered to simultaneously bind two distinct targets, achieving 'dual-target synergy'[61,62]. Cocktail drugs are highly active antiretroviral therapy commonly used in the treatment of HIV. Effective combination antiretroviral therapy can durably suppress HIV viremia and has dramatically improved HIV-associated morbidity and mortality[63,64].
FDC is another treatment modality that contains two or more active ingredients in single-dosage form[65]. For instance, sulfamethoxazole inhibits bacterial dihydrofolate synthetase, while trimethoprim inhibits dihydrofolate reductase[66]. Their combination, Compound Sulfamethoxazole, dually blocks bacterial folate synthesis (folate is essential for bacterial nucleic acid synthesis), increasing antibacterial activity by dozens of times compared to single components and reducing the likelihood of drug resistance[67]. A third form of drug combination in clinical practice is herbal remedies or traditional Chinese medicine (TCM) compound prescriptions, which contain a group of ingredient combinations that interact with key nodes in the occurrence and development of diseases, thereby exerting overall coordinated therapeutic effects[68,69]. Taking Compound Danshen Dropping pills as an example, which has been proven to confer multiple cardiovascular benefits[70,71]. Among them, tanshinones activate the Nrf2 pathway through HIF-1α and PKC/PKB, thereby enhancing intracellular endogenous antioxidant capacity[72]. In addition, salvianolic acid components can inhibit cardiomyocyte apoptosis by neutralizing reactive oxygen species (ROS)[73−75], exerting a temporally synergistic effect with tanshinones.
Clearly, current combination drugs are largely limited to the random or empirical combination of existing drugs, and the indications of combination therapies have generally been confined to some types of malignancies and cardiovascular disease, with much to be explored in many other diseases[76]. Notably, a research paradigm for the development of combination drugs derived from de novo synergistic targets or targetome is considerably lacking, especially considering the limited target landscape worldwide. Therefore, delving into the etiology and pathogenesis of disease development, identifying key signal nodes with synergistic integration effects, and further discovering and confirming target combinations with synergistic effects, developing combination drugs with clear action targets and synergistic mechanisms, emerge as important development trends in the current research and development of next-generation combination drugs for chronic diseases[77].
Based on background and these trends, here a trilogy of modern combinatory drug research is proposed (Fig. 4). The first and prevailing route is the combination of marketed drugs, as exemplified by Entresto; an alternative approach is the combination of an original drug with marketed drugs, as currently pursued by a prospective phase 2 study of combination epigenetic therapy against relapsed/refractory peripheral T cell lymphoma (PTCL), which combined azacitidine with chidamide to treat PTCL[78]. Moreover, a third and underexplored strategy is the de novo screening and rational design of combination drugs driven by targetome identification, which could be defined as the next-generation therapeutics.
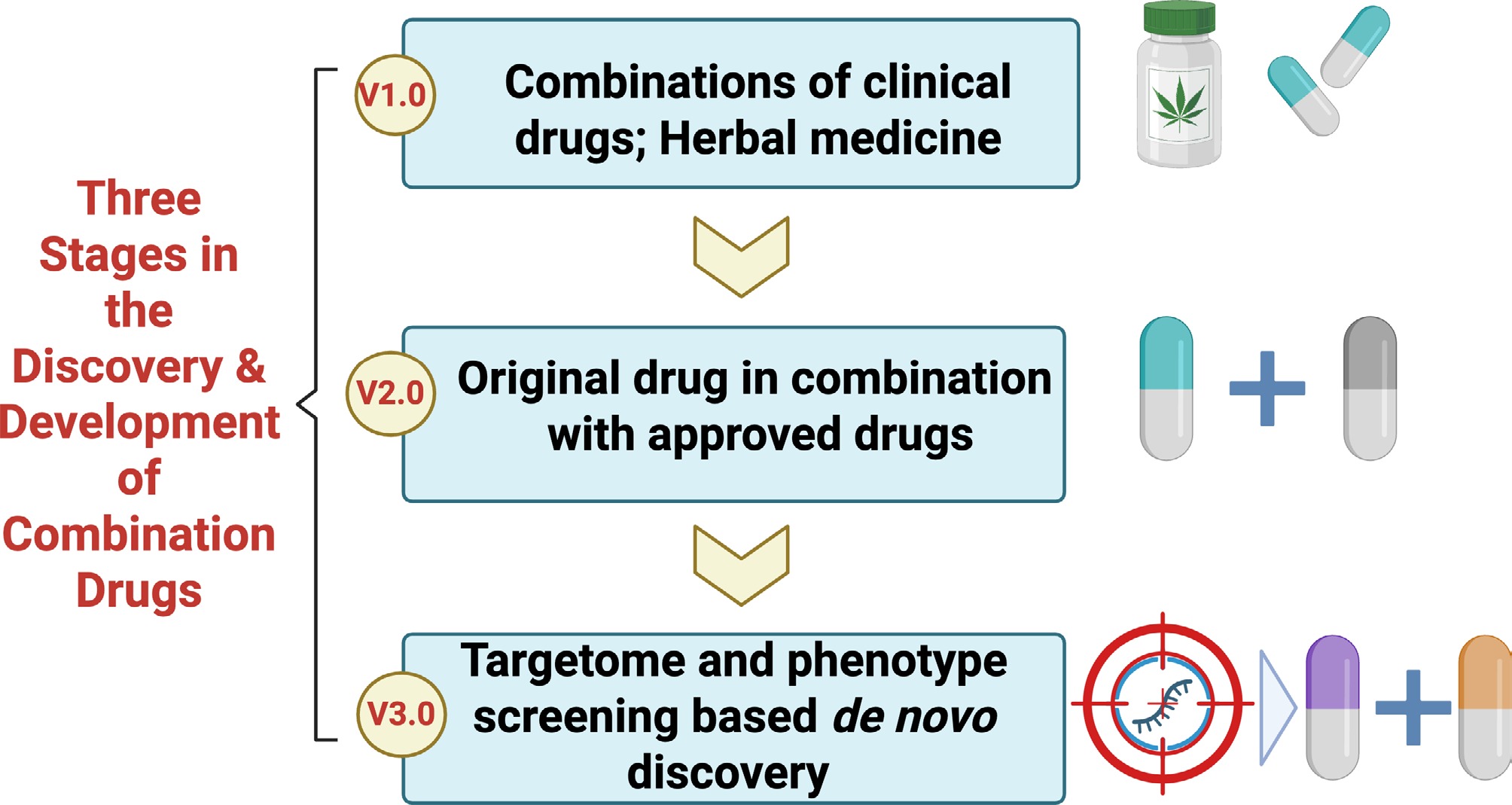
Figure 4.
Proposal of three stages in the discovery and development of combination drugs. Step 1: combinations of clinical drugs and herbal medicine are adopted for the exploration of combination therapies. Step 2: original drugs are combined with approved ones. Step 3: de novo discovery of combination drugs is carried out based on targetome and phenotype screening.
-
The transition from the 'one gene, one drug' paradigm to multi-target/node regulation is becoming popular, aiming to overcome the limitations of single targets through synergistic effects, ultimately enabling efficient treatment of complex diseases[79,80]. Common modalities for combination drug discovery are largely restricted to clinical experience-directed combination of old drugs. Here, from the perspective of de novo targetome identification validation, and drug design, a tentative framework is proposed for future endeavors in targetome-driven combination drug discovery.
Discovery of key signaling networks and synergistic targets for complex diseases
-
To explore the key signaling networks and synergistic targets in complex diseases, focus could be placed on basal biological processes such as cell death, immunometabolic and neural modulation, which constitute the central logic for the precise designing of combination therapies[81,82]. Abnormalities in cell death mechanisms are prevalent in essentially all chronic diseases and can be modulated through the combination of relevant drugs[83]; the crosstalk between metabolic and immune processes and the detection by the neural system have been implicated in many pathological conditions, which, for example, influences the tumor microenvironment and the progression of organ failure[84]. It is therefore reasonable that combination therapies can target key nodes in these core events to effectively ameliorate the complex diseases. A working route to the dissection of these key targets, as we proposed before, could be target identification of endogenous signaling molecules such as small-molecule metabolites and secreted proteins. Chemoproteomic approaches such as Target Responsive Accessibility Profiling (TRAP) technology[85], in conjudation with in vitro/in vivo functional validation and AI-based high-throughput analysis, may enable a rapid identification of the candidate targets. On this basis, multi-dimensional dissection of key signaling molecules and their spatiotemporal evolving patterns in major diseases, such as tumor, metabolic dysfunction, and cardiovascular diseases, represents the core of optimizing combination strategies. Ultimately, identifying the key molecular events that dictate disease progression and deciphering the dynamic regulatory patterns of signaling molecules are pivotal for achieving combinatory therapies[86,87].
Evaluation and validation of the druggability of combination targets
-
One of the core objectives of drug combination is to block disease compensatory pathways or enforce intervention efficacy by targeting combined targets within disease networks[88]. In the absence of a comprehensive evaluation on the properties of candidate targets, 'ineffective combination' or even 'antagonistic combination' may arise[89]. Thus, it is imperative to systematically validate the druggability of identified key signaling molecules and regulatory nodes from multiple dimensions, encompassing tissue-cell specificity, compensatory feedback, redundant pathways, synthesis-modification-degradation lifecycle, endogenous ligand-protein interactome, and the formation of signaling complexes. By assessing inter-target associations such as the crosstalk of combined target signaling pathways and gene co-expression patterns, combinations with genuine synergistic potential are screened to scientifically determine the potential druggability of synergistic targets, thereby offering theoretical guidance for drug design. For instance, in melanoma treatment, the combination of BRAF inhibitors (targeting BRAF V600E) and MEK inhibitors (targeting MEK1/2) has been evaluated to block upstream and downstream nodes of the MAPK pathway, respectively[90]. This combination can synergistically inhibit tumor proliferation and mitigate pathway rebound induced by single-agent therapy (e.g., MEK compensatory activation) and has ultimately received clinical approval (dabrafenib + trametinib).
Combination drug design for synergistic targets
-
Structural optimization and modification based on the discovery of endogenous ligands could become an efficient route. Endogenous ligands naturally occurring in organisms—such as hormones, neurotransmitters, and cytokines—are employed as prototypes. Through chemical modification approaches (e.g., adding/removing functional groups, altering configurations), their physicochemical properties are optimized or the side effects of natural ligands are attenuated, ultimately leading to the development of superior drug molecules[91,92]. The binding of these endogenous ligands to their targets exhibits inherent suitability, and drugs designed based on this framework demonstrate enhanced safety, with their mechanisms of action being more readily predictable[93]. Moreover, drug design based on three-dimensional structural analysis of targets is another fruitful route. Leveraging AI to predict the three-dimensional structure of target proteins, the binding modes between drug molecules and targets, or directly generate de novo candidate molecular structures has emerged as a prominent strategy in contemporary drug design. AI technologies not only process large-scale molecular datasets for the prediction of high-potential target combinations, but are also well-suited for designing multi-target combination drugs by predicting the binding capacity of molecules to multiple targets. This entails resolving the three-dimensional structure of target proteins via techniques like X-ray crystallography and cryo-electron microscopy, defining the spatial features of their active sites, and subsequently designing molecules that bind complementarily to these active sites to abrogate target function[94].
Innovative combined drug intervention could also derive from different drug modalities. For example, gene-cell therapy combinations entail modifying cells via gene editing or cell engineering technologies to enable simultaneous expression of therapeutic molecules targeting multiple targets, achieving precise and sustained disease intervention. The combination of CAR-T cells with IL-15 gene modification exemplifies this approach. Traditional CAR-T cells have short in vivo survival and are susceptible to suppression by the tumor microenvironment. Researchers introduced the IL-15 (a T cell survival-promoting cytokine) gene into CAR-T cells via lentiviruses, enabling them to secrete IL-15 autonomously while killing tumor cells (via CAR-mediated tumor antigen recognition), thereby extending their survival and enhancing activity. In refractory B-cell lymphoma, this combination therapy increased the complete response rate by over 30% compared to traditional CAR-T[95].
-
Attributed to the well-confirmed clinical benefits and safety profile, natural products serve as a vital source for combination drugs in light of the multi-component, multi-target working mechanism. Taking Compound Danshen Dropping Pills as an example, studies have identified an 'equivalent component group' consisting of 18 ingredients, which has demonstrated pharmacological effects comparable to those of Compound Danshen Dropping Pills in both cell models and rat models of myocardial infarction and ischemia-reperfusion[96,97]. The idea of efficacy-guided equivalent component discovery for TCM prescriptions thereby may open up a new path for the development of modern Chinese medicine and combinatory drugs[98], and a practical roadmap for trimming this complexity to a manageable 'modern equivalent component group' is needed.
However, TCM prescriptions—typically formulated with cultivated herbal materials—exhibit inherent batch-to-batch variability in metabolite profiles. Driven primarily by factors including herbal cultivation conditions (e.g., climate, soil properties, growth duration), such variability raises concerns regarding their eligibility for development as modern combination drugs. Addressing these concerns is essential to validate the reliability of TCM-based combination drug development. To mitigate the aforementioned batch-to-batch variability and enhance the translational potential of TCM prescriptions, emerging strategies may include: (i) integrating metabolomics with chemometrics to predict batch-specific biological activity, enabling pre-evaluation of pharmacological efficacy via metabolite profiles; (ii) employing multi-batch mixing to balance phytochemical fingerprints, thereby mitigating individual batch differences' impact on overall drug performance; and (iii) implementing bioassay-based standardization (e.g., NF-κB reporter gene) to establish consistent efficacy benchmarks aligned with modern pharmaceutical quality control standards.
Additionally, bioinformatics approaches matching natural products with potential targets are helpful to identify the druggable targets for novel combination drugs. For example, studies have combined single-cell multi-omics with target validation technologies to clarify that lobeline can directly target MAPK14, thereby regulating the p53/Slurp1 signaling pathway to promote the transformation of tumor-associated macrophages (TAMs) into an anti-tumor phenotype and inhibit the growth of colorectal cancer[99].
Endogenous molecules provide sources and inspirations
-
Endogenous molecules like small-molecule metabolites and peptides are bioactive compounds naturally produced in organisms, which are involved in numerous physiological processes in the body. Many endogenous molecules (e.g., ursodeoxycholic acid, insulin) have been approved as clinical therapies, and their disturbances have been implicated in many chronic diseases. As previously summarized by the present authors[42], a salient feature of many endogenous metabolites is the engagement with multiple targets to produce versatile effects across multiple cells/organs. The unique chemical skeletons and structure-bioactivity relationships could provide useful information and scaffolds for drug discovery through structural modification and optimization. For example, fibroblast growth factor 21 (FGF21)-based analogs prove effective to improve MASH, and a recent study attributes this benefit to brain and liver dual mechanisms that lower hepatic triglyceride and cholesterol levels[100].
The multi-metabolite multi-sensor/receptor working model of endogenous molecules is in line with the logic of targetome regulation. Previously, we have proposed a concept of metabolic bionics for the discovery of new drugs inspired by the action mode of endogenous metabolites, or bioactive signals[42]. Specifically, it focuses on the core wisdom of the biological system—inner homeostasis is maintained through multi-layer balanced regulation of the signaling network, ultimately aiming to restore, repair, or enhance the body's own rehabilitation. Of particular interest, hyocholic acid (HCA) was recently reported to stimulate GLP-1 production and secretion in enteroendocrine cells (EECs) via simultaneously activating TGR5 and inhibiting FXR. Such a unique mechanism indicates that promiscuous ligand-receptor interaction may be physiologically exploited for precision and balanced control of signaling transduction[42,101]. In this sense, studies on targetome identification and validation could gain inspiration from the working mode of endogenous molecules, where agonist/antagonist balance on a single target or signaling pathway increases the druggability. In analgesic research, for instance, balancing μ-opioid receptor (MOR) agonists and σ1 receptor (σ1R) antagonists led to novel 1-oxa-4,9-diazaspiro(5.5)undecane derivatives, among which compound 15au exhibits balanced dual profiles (MOR agonism and σ1R antagonism) with potent analgesic activity, offering insights for identifying endogenous metabolites or analogs with similar effects[102].
How to design such multi-target and multi-functional drugs has always been a challenge. Notably, a new multi-target and multifunctional drug design concept called 'Flexible Skeleton based Chemical Informatics Method (FSCA)' has been proposed. This is achieved by comprehensively applying multidisciplinary techniques such as chemical informatics, structural biology, and in vivo animal behavior[103]. Further application of this method to targetome regulation provides a new path for multi-target drug development and the treatment of complex disorders.
Multi-dimensional regulation of receptor/signaling networks
-
Drug targets exist in a dynamic status where their expression and activity are subjected to multi-dimensional regulation such as post-translational modification, co-regulator interaction, signaling complex aggregation, and degradation. Therefore, in-depth elucidation of the target regulation rules within the disease network provides the basis and targets for the design of combinatory drugs.
Designing of target stabilizer/degrader, for example, could serve as such an approach. A massive decrease in FXR was observed under hepatic diseases such as MASH. By elucidating the mechanism of FXR degradation, we propose that traditional FXR agonists, which encounter failure for MASH treatment, should be fostered by FXR stabilizers such as SUMOlyzation inhibitors to overcome the clinical dilemma[104]. Developing such drugs requires in-depth analysis of target dynamics within the signaling network. For example, cyclic GMP-AMP (cGAMP) is a second messenger that activates the stimulator of interferon genes (STING) innate immune pathway to induce the expression of type I IFNs and other cytokines. Podofilox, a microtubule destabilizer acting as a STING agonist enhancer, revealed interactions between microtubule systems and the STING pathway, opening new avenues for multi-dimensional regulation of STING, and related immune networks[105].
Multi-dimensional regulation of signaling networks also provides a fresh approach to augment treatment responses and clinical benefits, as exemplified by a recent study showing that ketamine-induced CA3-CA1 synaptic potentiation could be augmented by transiently increasing extracellular signal-regulated kinase (ERK) activity through pharmacological inhibition of dual-specificity phosphatases 6 (DUSP6)[106]. The combinatorial use of a DUSP6 inhibitor could therefore be rationally designed to prolong the antidepressant efficacy of ketamine.
Drug repurposing-guided combinatory drug discovery
-
Drug repurposing, the application of approved drugs for new indications, provides an efficient path for the findings of new treatment modality[107]. In the discovery of combination therapies, the value of drug repurposing is manifested in multiple aspects. On one hand, the pharmacokinetic, toxicological, and other relevant data of marketed drugs are relatively well-characterized, enabling faster evaluation of the safety and feasibility of combination regimens[108]. On the other hand, drug repurposing can uncover previously unrecognized pharmacological effects of drugs, providing novel strategies for combination therapy development[109]. For example, aspirin, initially used for antipyretic and analgesic purposes, was later found to possess antiplatelet aggregation activity[110]. In the treatment of cardiovascular diseases, it is often combined with clopidogrel to form a dual antiplatelet therapy regimen for thrombosis prevention. Furthermore, with the aid of modern information technology and bioinformatics analyses, candidate drugs can be more efficiently screened from marketed agents for combination therapy. A salient example is a recent study integrating single-cell transcriptomics, drug perturbation databases, and clinical records for drug repurposing. Using this framework, letrozole and irinotecan were identified as a potential combination therapy for Alzheimer's disease (AD), each targeting AD-related gene expression changes in neurons and glial cells, respectively[111].
AI and big data guided prediction of functional signal nodes in synergy
-
Integration of AI with pharmaceutical sciences holds profound promise for multi-drug combination strategies. In traditional research, identifying synergistic nodes often relies on extensive experimental trial-and-error, which is inefficient and time-consuming. The evolvement of AI models, however, enables the integration and analysis of massive biological datasets, thereby accurately predicting potential synergistic signaling nodes and guiding the development of combination therapies. In particular, harnessing the robust data processing and analytical capabilities of AI technologies, association networks between key signaling molecules and disease phenotypes can be constructed. Through causal prediction, key signaling molecules that may exert synergistic effects under combination drug interventions are identified[112]. Subsequently, the intricate regulatory mechanisms of these molecules are elucidated, and key signaling molecules along with their upstream and downstream pathways are translated into quantifiable mathematical models. Based on such models, the regulatory effects of combined targets under drug co-administration are theoretically predicted, followed by experimental validation of these predictions to confirm the authenticity and reliability of the observed effects[113].
Indeed, AI models and big data are empowering targetome research across the entire chain of 'target discovery-network mechanism analysis-molecular design', significantly improving the efficiency and accuracy of drug discovery. In terms of synergistic network modeling, the complexity of cellular signaling networks makes single-target intervention vulnerable to compensation mechanisms. AI technology can effectively mine key signaling nodes and combine intervention strategies by integrating multi-omics data. For example, the Geneformer model leverages transfer learning to learn the dynamics of gene networks from 30 million single-cell datasets. Even in rare disease research with limited data, it can accurately predict key regulatory factors and therapeutic targets in fields such as myocardial diseases[114]. At the de novo drug design level, AI models break through the limitations of natural molecules and enable entirely new designs at the molecular and protein levels. For example, an AI platform developed by the MIT team can generate 29 million novel antibacterial molecules, which effectively kill drug-resistant bacteria in mouse models. This validates the potential of AI in opening up new chemical spaces and overcoming drug resistance[115].
The aforementioned technologies therefore, formed a complete workflow in practical research. Taking the study on the synergistic regulation of the EGFR signaling pathway in non-small cell lung cancer (NSCLC) as an example: first, multi-omics data (genomics, transcriptomics, and proteomics) and drug perturbation data from patients are collected; second, AI models (such as DeepMind's deep learning models) are used to analyze signaling networks and identify potential synergistic nodes (e.g., PIK3CA and BRAF); third, a mathematical model of the signaling pathway is constructed to predict the impact of combined drug administration on network homeostasis; finally, the effectiveness of the scheme is verified through in vitro cellular experiments and in vivo animal models[116]. This closed-loop workflow greatly improves the success rate and translation speed of combination therapy development.
Phenotype screening-based discovery of combinatorial drugs
-
As early as the last century, phenotypic screening played a pivotal role in the discovery of many drugs, such as Chlorpromazine and Ibuprofen. Recent years have also seen a renaissance of this method in drug discovery[117]. Early phenotypic screening, using cell lines and defined compound testing methods, identified over 1,000 natural compounds with antibacterial and antifungal activity by observing cell inhibition or death without prior knowledge of mechanisms[118]. Nowadays, advanced technologies, including high-content automated imaging to capture drug-induced cellular morphological features for AI-driven analysis and machine learning to parse relationships in integrated multi-parameter phenotypic data, are useful to link cellular phenotypes to modes of action.
With technological advancements, next-generation phenotype-based screening offers greater advantages in multi-drug combination research. Combination screening models can simulate interactions between different drugs under diverse disease states; for example, when studying combination therapies for diabetes mellitus complicated with cardiovascular diseases, constructing combination models involving multiple cell types and signaling pathways enables a more comprehensive reflection of drug effects. Composite indicators break the limitations of single indicators by integrating metrics such as blood glucose control, improved vascular function, and reduced inflammatory responses to assess the efficacy of combination therapies[49]. Cutting-edge biotechnologies like cell painting, through high-content analysis of cell morphology, can rapidly identify the effects of different drug combinations on cells, providing rich phenotypic data for multi-drug combination screening. Integrating spatiotemporal omics as readouts allows for interpretation of dynamic changes in molecular phenotypes (e.g., gene expression, protein alterations) under combined drug action across temporal and spatial dimensions. For instance, in studying anti-tumor combination drugs, spatiotemporal omics can reveal how drugs affect the tumor microenvironment in different tissues and at different time points, thereby optimizing combination regimens.
In the screening of combination drugs, high-content and precision model design based on key signaling nodes is a critical strategy. The pathogenesis of many diseases is associated with abnormalities in key signaling pathways; designing combination therapies targeting these nodes can enhance therapeutic efficacy. In oncology, for example, the EGFR and PI3K-Akt-mTOR pathways are key nodes[119]. Combined use of EGFR inhibitors and mTOR inhibitors can synergistically suppress these pathways, enhancing anti-tumor effects—a finding supported by numerous studies. Maintaining metabolic homeostasis is fundamental to normal bodily function; multi-drug combinations must consider their impact on metabolic homeostasis, with screening focused on drug pairs that synergistically regulate metabolic disorders. For instance, in treating obesity-related metabolic syndrome, combinations that both improve insulin resistance and regulate lipid metabolism are prioritized[120]. Intercellular communication and organ crosstalk play important roles in disease progression, so screening strategies must focus on how drug combinations influence these interactions. In studying combination therapies for hepatopulmonary syndrome, for example, attention is paid to how drugs regulate hepatocyte-pulmonary cell communication and liver-lung crosstalk to restore normal organ functional interactions.
Targeting the host-microbiota interface and crosstalk
-
With the rapid progress of microbiota research, especially those inhabiting the gut, combination drugs targeting the host-microbiota interface can find potential sources by regulating microbiota composition and function, as well as modulating host immunity or physiological functions[121]. In addition to prebiotics and probiotics, which target the microbes, potential sources and strategies for host-targeted combination drugs include agents modulating host immune responses and those affecting host metabolism[122]. Statins, with anti-inflammatory and immunomodulatory properties, enhance host defense against pathogens by inhibiting the mammalian target of rapamycin (mTOR) pathway to promote autophagy and phagosome maturation—emerging as potential combination therapies for tuberculosis, focusing on pathogen-host immune cell interactions[123]. Another study has, for the first time, clarified the potential role of Fusobacterium nucleatum in regulating anti-PD-1 immunotherapeutic responses in microsatellite-stable colorectal cancer and its molecular mechanism. This study provides new ideas and strategies for the immunotherapy of patients with microsatellite-stable colorectal cancer, and also offers a new direction for anti-tumor immunity research[124].
-
The rapid advancement of combinatory drug research has provided a promising therapeutic paradigm across multiple diseases; yet, multifaceted scientific and technical challenges have also emerged in successfully translating to clinical benefits. How to get the right combination for the right person/patients is another open question. In the following section, we briefly discuss the open questions and bottlenecks that require theoretical and technical innovations.
Precision regulatory principles of targets
-
Targetome research aims to systematically identify and analyze all disease-modifying targets, thereby necessitating a more comprehensive understanding of their interrelationships with signaling networks. Signaling proteins and other biologic molecules are tightly and dynamically regulated in a full lifetime manner, broadly involving epigenetic and translational regulation, posttranslational modification, and protein-protein interactions, which critically affect the location, function, and destiny of candidate targets[125]. Therefore, targetome identification and combinatorial drug design should pay attention to such precision regulatory principles by employing various approaches, such as integrating omics platforms with high-throughput screening technologies to systematically identify synergistic nodes for phenotype regulation.
Spatiotemporal heterogeneity and integration of signals are essential aspects of biological signal regulation. Spatiotemporal heterogeneity refers to the variation in biological signals across different spatial locations and over time. Signals can vary in intensity, frequency, duration, and distribution within cells, tissues, and organs, allowing for precise and context-dependent responses. In the discovery of target combination for chronic diseases, it is therefore critical to understand the mechanism linking spatiotemporal signal integration to pathophysiology phenotypes and the treatment responses[126]. As a typical example, JAK1 is a key protein in the human body that supports cell communication and controls the immune system[127]. It is part of a group of proteins that transmit signals from outside the cell to inside the cell. Regulating JAK1 activity is important for the treatment of diseases such as rheumatoid arthritis and certain cancers, and current JAK inhibitors work well in reducing inflammatory responses in diseases like eczema[128]. The study indicates that JAK activity in pulmonary neurons may need to be enhanced rather than blocked, which could be a transformative strategy, different from traditional JAK inhibitors that mainly target immune cells. Subsequently, a mouse carrying a specific mutation in the JAK1 gene was used to reveal how this mutant protein causes diseases and how it can be used for broader therapeutic purposes[129]. These findings show that activated JAK1 signals have tissue-specific effects, including an unexpected immunomodulatory role in pulmonary sensory neurons, which inhibits lung inflammation. This explains why JAK1-selective inhibitors, despite being highly successful in atopic dermatitis, have made no progress in asthma treatment, meaning that JAK1 signals have different, even opposite, effects in different cell types and tissues. This finding also carries implications for therapeutic treatment. For example, during the initial phase of EGFR-TKI therapy or upon detection of MET amplification, combination with MET inhibitors (e.g., capmatinib) enables a timely blockade of signal transduction switching in the temporal dimension, thereby delaying the emergence of drug resistance. Clinical investigations have demonstrated that such a combinatorial strategy can almost double the progression-free survival in patients with drug resistance induced by MET amplification. Together, these findings underscore the need to better interpret signaling crosstalk and integration at spatial (subcellular, intercellular, and cross-organ), and temporal (initiation, progression) levels[130].
Validation of the targetome and clinical translation
-
At the level of target validation and confirmation, how to construct a theoretical and evaluation system to improve the success rate of clinical trials is a key challenge that needs to be overcome[131]. It is important to gain a comprehensive understanding of clinical relevance based on a human disease database, while appreciating the inherent differences in the biology of different diseases and disease phenotypes. Inherent biological differences exist between different diseases and distinct phenotypes of the same disease[132]—for instance, Parkinson's disease patients are classified into tremor-dominant (TD), postural instability and gait difficulty-dominant (PIGD), and mixed (Mix) subtypes based on motor phenotypes, with varying pathological mechanisms and gene expressions among subtypes, including 30 significantly up- or down-regulated genes in the Mix subtype, necessitating the process for subtype-specific valiations[133].
The clinical evaluation of combination drugs needs to superimpose multiple layers of complexity on the traditional trial framework and achieve scientific rigor through dose optimization, dynamic monitoring, and precision stratification design. Regulatory policies, on the other hand, need to focus on technological innovation and reconstruct the evaluation paradigm through tools such as MBDD, AI-based review, and VCT, while strengthening the whole-life-cycle risk management[134,135]. In the future, with the integration of gene-editing technologies (e.g., CRISPR) and cell therapies, regulatory authorities may need to further explore the approval pathways for 'living drug' combinations, such as the combined therapy of CAR-T cells and immune checkpoint inhibitors, which will pose new challenges to the existing regulatory framework[136,137].
Pharmacokinetics and drug delivery considerations
-
At the level of drug design and combination drug discovery, how to ensure synergistic efficacy by addressing pharmacokinetic compatability should also be considered. AI Modeling to predict in vivo drug concentration-time curves of the individual component may guide dosage design, optimizing drug design to better align with the spatiotemporal requirements for target engagement[138]. Moreover, from the perspective of drug delivery, drug carriers could be designed by simulating the natural environment of targets and the in vivo transport mechanisms of natural nanocarriers, such as a 'cell-matrix-mimetic' biomimetic nanocarrier[139]. Future progress in smart drug delivery systems is expected to find extensive use in driving the clinical application of combination therapies.
-
Previous drug development strategies focusing on a single target or node have often demonstrated limited efficacy. In contrast, combination therapies that target two or more pathways have shown superior pharmacological activity on chronic diseases, raising a new direction in breaking the stalemate of chronic disease management. In this review, major progress in this frontier is synthesized and three potential routes proposed for discovering next-generation combination drugs. Key dimensions for targetome identification and validation are considered, based on the underlying pathophysiology and regulatory principles of disease and opportunities discussed to harness technical advances in disease biology and drug design to overcome challenges associated with these combinatorial strategies. Overall, AI and big data are driving targetome research from static map drawing to dynamic synergistic network analysis, and rational molecular design. In the future, with the development of multi-modal data and interpretable AI technologies, this model is expected to enable precise and efficient drug discovery and therapeutic strategy development in more disease fields.
-
Not applicable.
-
The authors confirm their contributions to the paper as follows: conceptualization: Hao H, Zheng X; writing of this manuscript: Hao H, Liu P, Huang F; revision of this manuscript: Zheng X, Liu P, Huang F. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
-
This work was supported by the National Key Research and Development Program of China (grant 2021YFA1301300 to H.H.), and the Natural Science Foundation of Jiangsu Province (grant BK20240095 to X.Z.).
-
The authors declare that they have no conflict of interest.
-
#Authors contributed equally: Pan Liu, Fuyue Huang
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press on behalf of China Pharmaceutical University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Liu P, Huang F, Zheng X, Hao H. 2025. Targetome-guided combination drug discovery as next-generation therapeutics. Targetome 1(1): e002 doi: 10.48130/targetome-0025-0002
Targetome-guided combination drug discovery as next-generation therapeutics
- Received: 05 September 2025
- Revised: 27 September 2025
- Accepted: 09 October 2025
- Published online: 22 October 2025
Abstract: The limited efficacy of 'one gene, one target' drugs for major chronic diseases such as cancer and cardiovascular disease has spurred the clinical development of combination therapies, with the hope of attaining synergistic activity and/or overcoming treatment resistance. This paradigm shift is witnessed against the accumulation of insights into the inherent complexity of biological systems and chronic diseases, which serves as the basic logic for the quest for drug combinations. Several combinatorial agents have been approved by the FDA, and many candidates are currently under clinical trial. However, the design of these drugs has been mostly restricted to the empirical combination of marketed drugs. Given the limited target landscape in the current drug development pipeline, there is an unmet need to identify innovative combination drugs based on de novo targetome discovery. In this review, major progress in this frontier is synthesized and three potential routes are proposed that could be pursued for the discovery of next-generation combination drugs. Key dimensions for targetome identification and validation are considered, based on the signaling network of chronic diseases and regulatory principles of targets. Opportunities for harnessing the technical advances in disease biology and drug design to overcome challenges associated with these combinatorial strategies are discussed.
-
Key words:
- Drug discovery /
- Combination drug /
- Targetome /
- Chronic diseases /
- Target identification