-
Peanut allergies are among the most common and serious food allergies, particularly in Western countries, affecting approximately 1%−2% of the population[1]. These allergies can trigger life-threatening reactions, such as anaphylaxis, which significantly affect individuals and their families due to dietary restrictions and social limitations. The main concern for individuals with peanut allergies is the fear of accidental peanut consumption. The prevalence of peanut allergies has been rising, particularly among children, prompting further investigation into environmental, immunological, and genetic factors that contribute to allergic reactions[2,3]. In regions such as North America and the United Kingdom, the prevalence of peanut allergies among school-aged children has surpassed 1%, which has raised public health concerns and calls for more research[4].
The avoidance of peanuts is essential for those affected, as it significantly influences their diet, social activities, and the food industry[5,6]. The primary proteins in peanuts responsible for triggering allergic reactions are Arah1, Arah2, and Arah3[7,8]. These allergens provoke immune responses that lead to symptoms like hives, swelling, respiratory distress, and even anaphylaxis. Arah2, in particular, is considered one of the most potent allergens[9]. Reducing peanut allergenicity presents a challenge, as modifying or removing these proteins without affecting key peanut traits such as nutritional value or taste is complex. Enzymatic hydrolysis has been identified as a potential method for reducing allergenicity; however, chemical modifications may also alter the properties of peanuts, potentially making them more allergenic[10,11].
Recent advancements in the development of allergen-free peanuts have been achieved through both traditional breeding and cutting-edge gene-editing techniques[12]. Traditional breeding efforts have focused on identifying natural peanut varieties with lower allergenic protein content and crossbreeding them to produce cultivars with reduced allergenicity[11]. The terms 'allergen-free' and 'allergen-reduced' are important in this context. 'Allergen-free' refers to the complete elimination of allergenic proteins, while 'allergen-reduced' means that the quantity or activity of these proteins has been decreased, though they may still be present at lower levels[11,13].
Epigenetic technologies offer promising strategies for controlling the expression of specific allergenic proteins, such as Arah1, Arah2, and Arah3[14]. Using tools like CRISPR-based systems, researchers can selectively downregulate the genes responsible for producing these allergens without altering the overall genetic makeup of the peanut[15,16]. This technique involves modifying DNA methylation or histone modifications at specific loci responsible for allergen expression[17,18]. Such a targeted approach could reduce peanut allergenicity, leading to peanuts that are safer for individuals with peanut allergies. Practical examples include using epigenetic tools, such as CRISPR-dCas9, to regulate allergen expression in peanuts.
In addition to Arah1, Arah2, and Arah3, Arah6 and Arah8 are also significant contributors to peanut allergies[19−21]. Arah6 is a 2S albumin, while Arah8 is a profilin, both of which are known allergens, particularly in individuals with latex-fruit syndrome, where there is cross-reactivity between latex and peanut proteins[22]. Epigenetic tools like CRISPR-dCas9 could be used to target and regulate the expression of these allergens. By modifying the expression of Arah6 and Arah8, it is possible to create peanuts with a reduced allergenic profile, offering safer options for individuals who are sensitive to multiple allergens[19,23].
Research on the epigenetic regulation of Arah6 and Arah8 is still in its early stages, but initial results are promising. Modifying the expression of these allergens without altering the rest of the peanut genome could lead to peanuts that are safer for individuals with specific sensitivities. Epigenetic silencing of Arah6 and Arah8 could help develop allergen-reduced peanuts that retain vital agronomic traits, such as yield and nutritional value while reducing the risk of allergic reactions[24].
Despite these advances, challenges persist in developing allergen-reduced peanuts. Limited genetic variability in peanuts and the complexity of allergenic protein profiles remain significant obstacles[25,26]. However, breeding programs have successfully produced peanut lines with reduced allergen levels, offering safer options for individuals with mild peanut sensitivities. Gene-editing technologies, particularly CRISPR-Cas9, have revolutionized the development of allergen-free peanuts by enabling precise modifications to the genes responsible for allergenic proteins[15]. Studies have demonstrated the successful silencing of genes such as Arah2 and Arah6, leading to a significant reduction in allergen levels[27]. This approach accelerates the development process compared to traditional breeding techniques while maintaining specificity and minimizing unintended effects on other peanut traits like yield and flavor.
Furthermore, advancements in molecular biology have enabled the combination of multiple allergen-reducing edits, potentially creating peanuts that are almost completely free of major allergenic proteins[10]. Although regulatory and public acceptance challenges remain, gene-edited peanuts have shown promise in preclinical studies, with allergenicity tests indicating their potential to be safer for consumption[28]. These innovations, along with ongoing traditional breeding efforts, offer hope for the widespread adoption of allergen-free peanuts, contributing to safer food systems and addressing the public health burden of peanut allergies.
While significant progress has been made in food science, the management of peanut allergies still relies primarily on avoidance and emergency treatment with epinephrine[29]. A review of peanut allergy management notes that while avoidance and the use of an epinephrine auto-injector are standard practices, the risk of fatal anaphylaxis or harm from skin contact or inhalation is relatively low[30]. Research into immunotherapy and desensitization techniques is still in progress, but a definitive solution has not yet been found. The main challenge remains the difficulty of removing or altering allergenic proteins like Arah1, Arah2, and Arah3 without compromising the peanut's nutritional quality or consumer acceptance. Thus, this review aims to explore epigenetic approaches for producing allergen-free peanuts, offering new possibilities for managing peanut allergies and enhancing food safety.
-
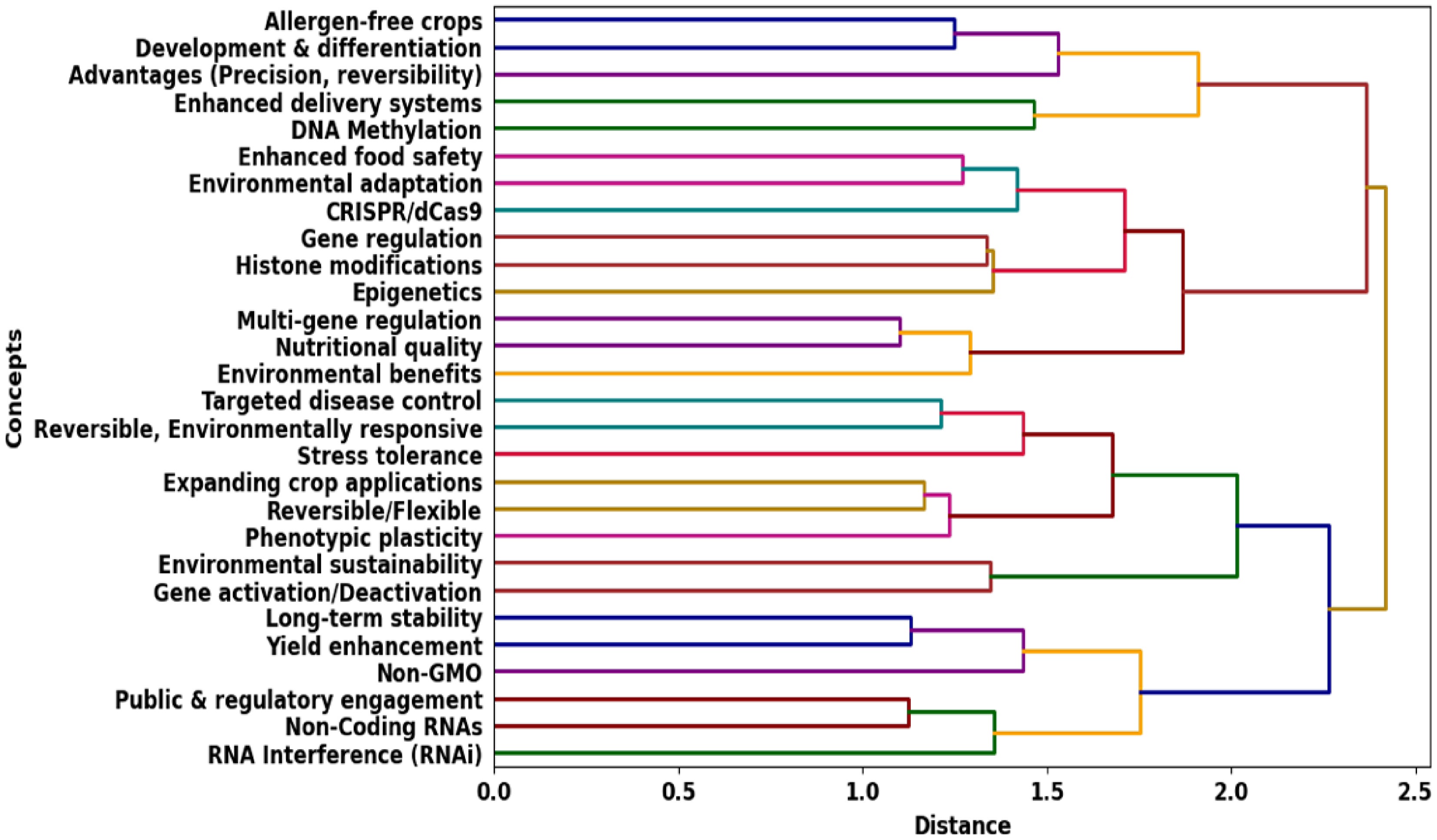
Epigenetics involves the examination of inheritable changes in gene expression that do not involve modifications to the DNA sequence, driven by mechanisms like DNA methylation, histone alterations, and non-coding RNAs[31]. In contrast to genetic mutations, epigenetic changes are reversible and can be shaped by environmental factors. Key processes in epigenetics include DNA methylation (which silences gene expression), histone modifications (which modify chromatin structure), and non-coding RNAs (which regulate gene activity after transcription)[32]. Epigenetic mechanisms offer flexible regulation of gene expression, which is essential for processes like development, cellular differentiation, and responses to environmental changes. A review study on precision epigenetic editing: Technological advances, enduring challenges, and therapeutic applications (human studies) states that the epigenome is a complex framework through which gene expression is precisely and flexibly modulated to incorporate heritable memory and responses to environmental stimuli[33]. A review of the fundamentals of epigenetics suggests that environmental factors can induce lasting phenotypic changes by causing heritable epigenetic alterations. These modifications influence nucleosomal organization and control whether genes are silenced or activated[34,35]. Likewise, epigenetics offers the potential to improve crop traits such as stress resistance, disease resilience, and yield by adjusting epigenetic markers, all without altering the plant's DNA. This provides a versatile tool for adapting crops to environmental challenges. Epigenetic editing tools, such as CRISPR/dCas9 and RNA interference, are transforming plant disease treatment and agricultural biotechnology by allowing precise control of gene expression without altering the underlying DNA sequence[36]. In plant disease management, these technologies can deactivate genes that increase susceptibility to pathogens or activate those that improve disease resistance, providing a targeted and sustainable method for reducing crop losses[37,38]. In agricultural biotechnology, epigenetic editing enhances traits such as stress tolerance, yield, and nutritional quality while also reducing allergenic proteins in crops like peanuts, making them safer for consumption[39]. This precise regulation of gene activity minimizes unintended effects, offering innovative solutions for managing plant diseases, improving food safety, and promoting agricultural sustainability.
The dendrogram (Fig. 1) illustrates the hierarchical relationships between key epigenetic concepts and their agricultural applications. It emphasizes the connections between genetic regulation, plant disease management, and sustainable farming practices.
-
Epigenetic editing tools are molecular technologies that precisely modify epigenetic marks, such as DNA methylation and histone modifications, to regulate gene expression without changing the DNA sequence[40,41].
Epigenetic editing technologies offer innovative possibilities for creating allergen-free peanuts by accurately regulating gene expression without modifying the DNA sequence itself[42]. A book chapter on CRISPR-Cas technology emphasizes that this system offers several advantages over earlier gene-targeting methods like Transcription Activator-Like Effector Nucleases (TALEN) and Zinc Finger Nucleases (ZFN). These advantages include simpler design, precise genome editing in living cells, higher efficiency, and lower costs[43]. A promising approach is CRISPR/dCas9-based epigenetic editing, which, unlike conventional CRISPR, does not cut DNA but instead silences specific allergen genes such as Arah1, Arah2, and Arah3 through epigenetic changes, thereby reducing allergen production[44].
RNA interference technology effectively reduces the expression of allergenic proteins by targeting the mRNA of specific allergen genes, thereby decreasing allergen levels in peanuts. This approach has proven successful in other crops, indicating its potential for application in peanuts[45,46].
Additionally, emerging epigenetic tools, such as small molecules that modify histones or DNA methylation patterns, are being explored to reduce allergens[47]. These methods enhance the toolkit for developing hypoallergenic peanut varieties and represent cutting-edge strategies for addressing peanut allergies through epigenetic modification[48]. Epigenetic editing technologies, such as CRISPR/dCas9, RNA interference, and DNA methyltransferases, allow for precise and heritable suppression of allergenic genes without modifying the DNA sequence, presenting a promising approach for developing allergen-free crops (Table 1).
Table 1. Epigenetic editing tools that could be applied for allergen-free peanut production.
No. Epigenetic editing tools Description summary Ref. 1 CRISPR/dCas9 (Dead Cas9) Used for targeted gene repression by modifying gene expression without cutting DNA. [49] 2 Transcription activator Engineered to precisely target DNA sequences for the epigenetic regulation of gene expression. [50,51] 3 Zinc finger nucleases Engineered proteins that target and bind specific DNA sequences to induce epigenetic changes. [52] 4 RNA interference Utilizes small RNAs to silence allergenic gene expression at the post-transcriptional level. [53] 5 DNA Methyltransferase Used to target and methylate specific gene regions, silencing allergenic genes. [54] 6 Histone deacetylase Employed to alter histone acetylation, repressing allergenic gene expression by tightening chromatin. [55] 7 CRISPR/Cas13 Used to degrade allergenic transcripts in peanuts. [56] 8 Non-coding RNA-based tools Regulate the expression of allergenic genes through epigenetic mechanisms. [57] -
Peanut food allergy is one of the most severe and common food allergies, affecting millions worldwide[1]. It can trigger life-threatening reactions such as anaphylaxis, characterized by difficulty breathing, a rapid drop in blood pressure, and, in severe cases, death[58]. Even small amounts of peanut proteins can provoke allergic reactions in sensitive individuals, making this condition a significant public health concern[59,60]. Peanut allergy often starts in early childhood and tends to persist throughout life, significantly impacting dietary habits and quality of life[60].
To address peanut allergies, researchers have explored various methods to create allergen-free peanuts. One promising approach involves genetic engineering to silence or modify the genes responsible for producing allergenic proteins[61,62]. RNA interference (RNAi) technology has been used to suppress the expression of Arah2 and Arah3 proteins, significantly reducing allergenicity. Additionally, breeding techniques, such as traditional crossbreeding or CRISPR-Cas9 genome editing, are being employed to produce hypoallergenic peanut varieties[63]. Another method involves post-harvest processing, such as enzymatic treatments, to break down allergenic proteins[64]. These approaches hold great potential for providing safer peanuts while maintaining their nutritional and sensory properties.
Targeted epigenome editing is an advanced technique that enables the precise modification of epigenetic marks, such as DNA methylation or histone modifications, to regulate gene expression without altering the underlying DNA sequence[40,41]. For example, the review article on epigenome editing technologies for discovery and medicine reports that epigenome editing exploits the heritable and reversible mechanisms of epigenetics to alter gene expression without introducing DNA breaks, inducing DNA damage, or relying on DNA repair pathways[65]. The approach utilizes tools like CRISPR/dCas9, which pairs a deactivated Cas9 protein with specific epigenetic effector proteins to target specific genomic regions[66]. Researchers can regulate gene activity by adding or removing specific epigenetic marks, offering a flexible and reversible approach to controlling plant traits. This technique is particularly useful for managing traits governed by complex gene networks, such as improving stress tolerance, increasing yield, or enhancing resistance to diseases[36].
Applications of targeted epigenome editing in crops have shown immense potential. For example, in rice, researchers have modified epigenetic marks to improve drought and salt tolerance by targeting genes related to stress response pathways[67]. In tomatoes, epigenome editing has been used to extend shelf life by altering the expression of ripening-related genes[68,69]. Similarly, in maize, researchers have enhanced yield and biomass production by regulating key genes involved in growth and metabolism[70,71]. This technique offers an alternative to traditional genetic modification, addressing public concerns about permanent DNA changes while expanding the toolkit for sustainable agriculture. As research advances, epigenome editing could revolutionize crop improvement by offering precise, efficient, and environmentally friendly solutions[72].
The author of one study suggests that targeted epigenome editing could offer a novel and precise approach to developing allergen-free peanuts by regulating the expression of allergenic genes without altering the DNA sequence[42]. Tools such as CRISPR/dCas9 combined with epigenetic modifiers can be utilized to suppress the expression of allergen-encoding genes like Arah1, Arah2, and Arah3. This can be achieved by introducing repressive epigenetic marks, such as DNA methylation or histone deacetylation, at the promoters of these genes, thereby significantly reducing or completely silencing their transcription[6]. This would decrease the production of allergenic proteins, potentially creating peanuts that are hypoallergenic while maintaining their nutritional value and overall quality.
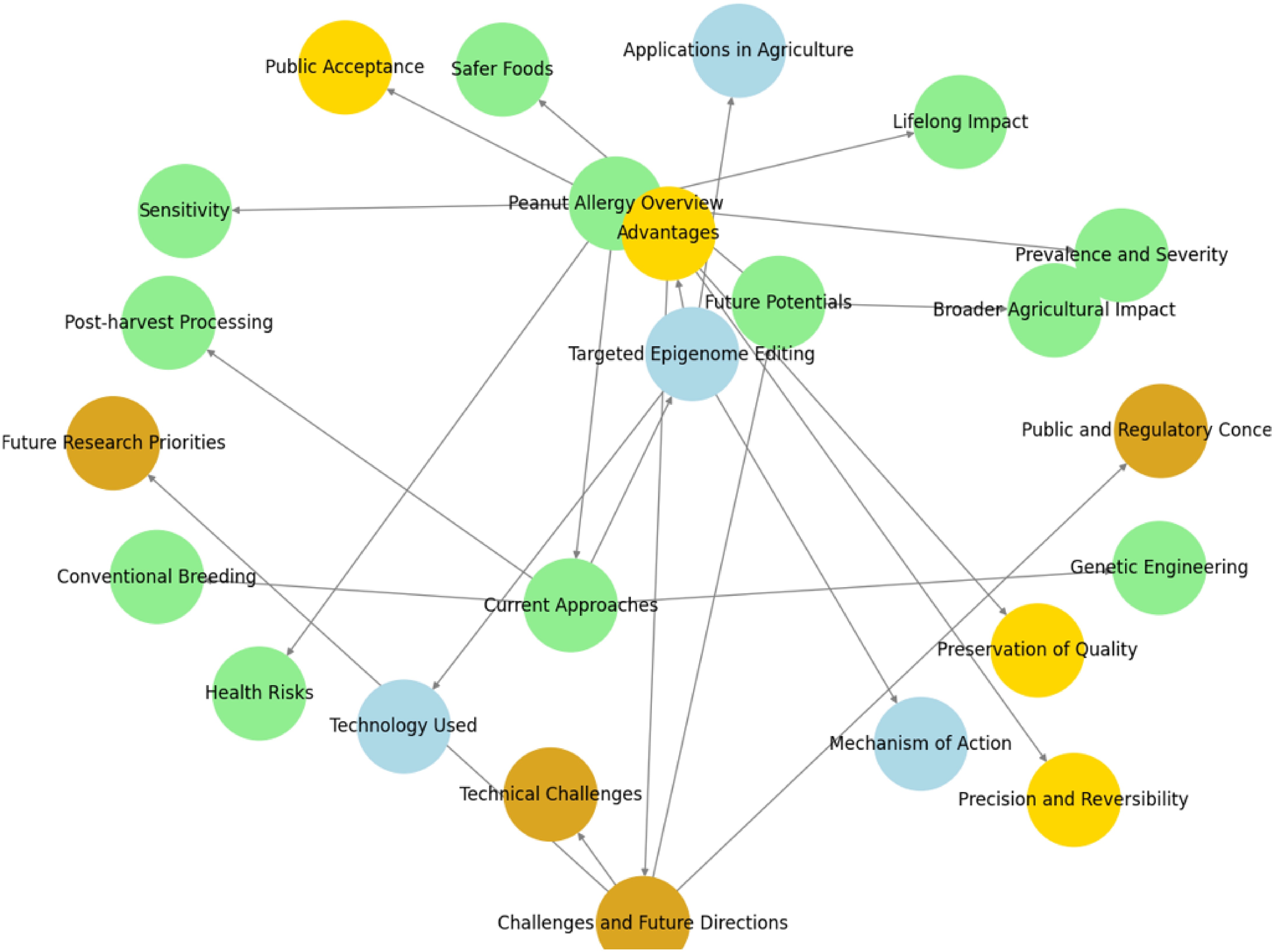
However, there are significant challenges to implementing epigenome editing for allergen-free peanut development (Fig. 2). A major issue is ensuring the long-term stability of the epigenetic modifications across multiple generations, as some marks may be reversible or lost over time[73]. Another challenge is achieving tissue-specific and precise targeting, as some allergenic proteins may have functional roles in peanut growth and development[39]. Additionally, off-target effects, where the editing machinery inadvertently affects non-target genes, must be minimized to prevent unintended consequences[74]. Regulatory and public acceptance of epigenetically modified crops also remains a significant hurdle, as such technologies are still relatively new in agriculture[75].
Looking ahead, future research should focus on optimizing the delivery systems for epigenome-editing tools, such as nanoparticles or plant transformation methods, to ensure efficient and precise gene targeting in peanuts[76]. Progress in understanding the epigenetic regulation of allergenic genes will be vital for increasing the consistency and reliability of these modifications. Additionally, combining epigenome editing with other breeding methods, such as CRISPR-Cas9 genome editing or RNA interference, could further improve the efficiency and success of developing allergen-free peanuts[43]. If these challenges are addressed, targeted epigenome editing could pave the way for safer, allergen-free peanuts and inspire broader applications in addressing food allergies in other crops.
-
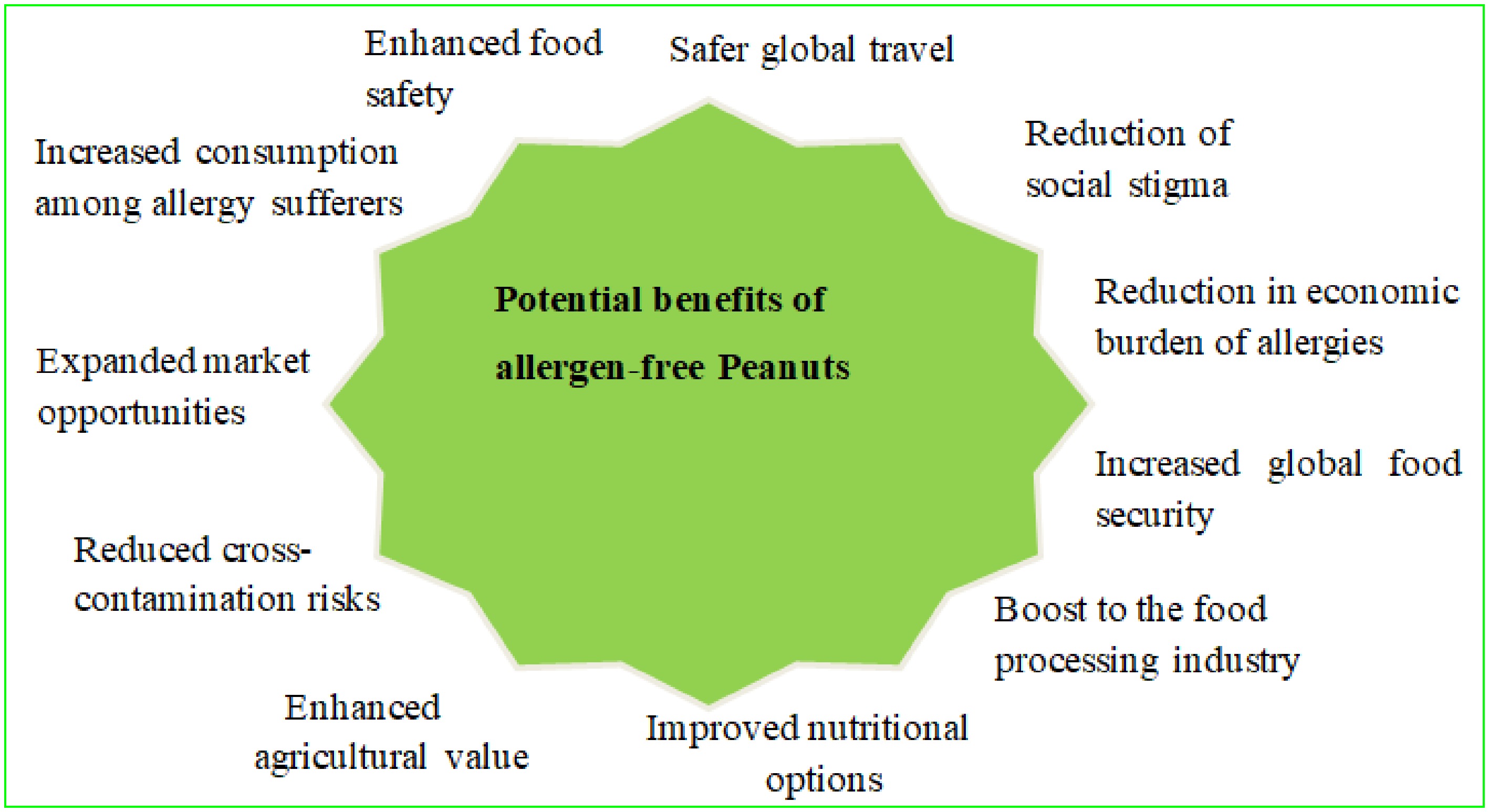
Allergen-free peanuts could have a significant public health impact by reducing the prevalence of peanut-related allergic reactions, including life-threatening anaphylaxis. A study on the risks of shared equipment in restaurants for individuals with peanut allergies, specifically in the context of preparing Asian dishes, concludes that an eliciting dose of 300 or 1,000 mg of peanut protein is clinically significant for the peanut-allergic population[77]. This innovation could greatly reduce the need for strict avoidance measures and emergency treatments for individuals with peanut allergies, improving their overall quality of life and potentially reducing the occurrence of allergies in future generations. The introduction of allergen-free peanuts may also support the growth of peanut-based products and open new market opportunities. A book chapter on the economic and academic significance of peanuts highlights the strong global demand for peanuts and peanut-based products, particularly in the confectionery sector[6]. Reducing concerns about allergic reactions would enable food manufacturers to include peanuts in a wider variety of products, unlocking new opportunities in the food industry[78]. Educational institutions, airlines, and other organizations could also safely reintroduce peanuts into their offerings, capitalizing on the peanut's versatility and nutritional value. The American Sociological Review article Another Person’s Peril: Peanut Allergy, Risk Perceptions, and Responsible Sociality suggests that the three dimensions of proximity to risk material, social, and situational play a key role in shaping policy discussions and influencing how people perceive fear, trust, rights, obligations, and freedom in shared environments, ultimately guiding their views on 'responsible sociality'[79]. Crucially, allergen-free peanuts would preserve their nutritional integrity, providing a sustainable source of protein, healthy fats, and essential nutrients. Research on the bioactive compounds in peanut skin for chronic non-communicable diseases highlights that, due to its polyphenolic content 60 to 120 times higher than that of the peanut grain peanut skin by-products serve as a sustainable source of plant-based bioactives beneficial to human health[80]. Hence, by mitigating allergens without compromising the peanut's nutritional profile, the peanuts could help to address the increasing global food demands while promoting environmental sustainability in peanut cultivation. Creating allergen-free peanuts holds great promise for improving public health, increasing market accessibility, and minimizing the risk of severe allergic reactions, all while fostering a more inclusive food industry (Fig. 3).
-
One of the major challenges in producing allergen-free peanuts through epigenetic approaches is achieving stable and consistent modifications. The book chapter 'Managing Food Allergens' proposes a method for controlling allergen risks by setting thresholds that trigger allergic reactions[81]. Ensuring consistent changes in allergenic gene expression, like Arah1, Arah2, and Arah3, throughout the peanut plant's lifecycle and across generations is a technical challenge, as variability could compromise efforts to reduce allergens.
Another challenge is comprehending the long-term effects and potential reversibility of epigenetic modifications. The Proceedings of the Nutrition Society on Nutritional Developmental Epigenomics: Immediate and Long-Lasting Effects suggests that epigenetic misprogramming during development is believed to have lasting impacts on offspring health, also potentially affecting subsequent generations[82]. Since epigenetic modifications do not change the DNA sequence, they can sometimes be reversed by environmental influences or during the plant's reproductive process. Hence, ensuring the stable inheritance of allergen-free traits without unintended effects is crucial for their agricultural viability[83,84].
Future research efforts should focus on creating more precise and durable epigenetic editing tools, with advancements in technologies such as CRISPR/dCas9 and RNA interference. It is equally important to evaluate the safety and efficiency of these modifications to ensure their viability. Additionally, investigating how environmental factors influence epigenetic alterations and fostering acceptance among consumers and regulatory bodies will be critical steps in advancing the production of allergen-free peanuts.
-
In conclusion, epigenetics offer a groundbreaking approach to developing allergen-free peanuts by precisely regulating the expression of allergenic genes such as Arah1, Arah2, and Arah3 without altering the underlying DNA sequence. Advanced tools like CRISPR/dCas9 and RNA interference allow for the effective suppression of allergenic protein production, presenting a non-GMO solution to peanut allergies. This innovative application of epigenetics could enhance food safety by reducing allergic reactions, improving the lives of those with peanut allergies, and potentially preventing allergies in future generations. Additionally, allergen-free peanuts could open new market opportunities by making peanuts acceptable in restricted environments such as schools and public spaces. This development would not only boost the peanut industry but also retain the crop’s nutritional value and environmental sustainability. Overall, epigenetic strategies have immense potential to revolutionize food safety, improve public health, and strengthen consumer confidence in hypoallergenic food options.
The authors would like acknowledge CAAS-ASTIP and CARS-MOF & MARA for providing the funding for this work. The Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2021- OCRI) and the China Agriculture Research System of MOF and MARA.
-
The authors confirm contribution to the paper as follows: draft manuscript preparation, review: Gelaye Y; study supervision, manuscript revision: Luo H. Both authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no new data were analyzed in this study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Gelaye Y, Luo H. 2025. Application of epigenetics for allergen-free peanut production: a comprehensive review. Epigenetics Insights 18: e006 doi: 10.48130/epi-0025-0006
Application of epigenetics for allergen-free peanut production: a comprehensive review
- Received: 08 October 2024
- Revised: 23 March 2025
- Accepted: 03 April 2025
- Published online: 13 May 2025
Abstract: Epigenetic methods provide a promising avenue for creating allergen-free peanuts, helping to address the increasing incidence of peanut allergies and related health issues globally. This paper thoroughly explores epigenetic approaches for producing allergen-free peanuts and offers new possibilities for effectively managing peanut allergies. By specifically targeting genes like Arah1, Arah2, and Arah3, the major allergens responsible for allergic reactions can be reduced. Innovative technologies such as CRISPR/dCas9 and RNA interference enable precise regulation of gene activity, facilitating allergen-free peanut production without altering the underlying DNA sequence. The advantages of allergen-free peanuts extend beyond the health benefits, as they have the potential to enhance food safety and open up entirely new markets for peanut-based products. These peanuts could be safely reintroduced into public spaces, like schools and daycare centers, increasing consumer trust and fulfilling the growing demand for safe, nutritious alternatives. However, challenges persist, such as ensuring stable epigenetic changes, preserving other essential peanut traits, and understanding their long-term effects on human health and the environment. Ongoing research should focus on refining these techniques to address these challenges, promote the successful integration of allergen-free peanuts into agricultural systems, and improve public health while transforming the food industry in a sustainable manner.
-
Key words:
- Allergen-free /
- CRISPR /
- Epigenetics /
- Gene expression /
- Peanut production