-
Nitrogen is fundamental to metabolism and a key factor in plant growth and yield[1]. As an indispensable mineral nutrient element for peach growth, nitrogen is key in regulating yield and quality. Elucidating key genes governing high nitrogen use efficiency in peach is critical for enhancing nitrogen utilization efficiency and reducing nitrogen fertilizer application in sustainable orchard management systems. Peach is a fruit tree that primarily absorbs nitrogen in the form of NO3−-N, using both low and high affinity transport systems[2]. At present, the understanding of the absorption and utilization of peach tree nitrogen is not comprehensive or specific enough. In agricultural practice, excessive nitrogen fertilization aimed at yield enhancement often results in 50%–70% of applied nitrogen remaining unutilized by crops[3]. This inefficient nutrient management not only incurs economic losses from fertilizer waste but also triggers a cascade of environmental consequences, including ecological degradation, soil quality deterioration, and nitrogen cycle disruption within agroecosystems[4,5]. Therefore, elucidating the molecular mechanisms underlying high-efficiency nitrogen utilization in peach, identifying nitrogen-efficient key genes, and enhancing NUE is critical for advancing sustainable production, and ecological sustainability in peach cultivation systems.
The root system, as a vital nutrient organ in plants, performs indispensable biological functions including plant anchorage, growth promotion, water/nutrient acquisition, and environmental adaptation. Its spatial expansion primarily relies on lateral root development mechanisms – specialized structures derived from epidermal cell differentiation that substantially increase root surface area, thereby enhancing nutrient and water capture efficiency[6]. During peach growth and development, the well-developed root system not only provides mechanical support for aerial growth but also significantly improves NUE through optimized nitrogen absorption and metabolic pathways. Plant nitrogen metabolism encompasses four key processes: absorption, assimilation, translocation, and remobilization[7]. The nitrogen assimilation process involves coordinated enzymatic mechanisms. In the initial phase of inorganic nitrogen conversion, nitrate reductase (NR), and nitrite reductase (NiR) constitute a dual-enzyme system for nitrate assimilation: NR catalyzes the reduction of nitrate (NO3−) to nitrite (NO2−), which is subsequently converted by NiR into ammonium (NH4+) or nitric oxide (NO) as intermediate products[8]. Notably, NiR serves as a pivotal nodal enzyme in the nitrogen metabolic network, with its catalytic activity simultaneously influencing both the assimilatory pathway (NH4+ production) and dissimilatory pathway (NO signaling molecule synthesis), playing a core regulatory role in maintaining nitrogen metabolic homeostasis[9]. Subsequently, the GS-GOGAT cycle system, formed by glutamine synthetase (GS) and glutamate synthase (GOGAT), governs the synthesis and translocation of organic nitrogen, ultimately completing the transformation from inorganic to organic nitrogen forms.
Transcription factors play a critical role in regulating plant growth and development. In the regulatory category of root development, many transcription factors (TFs) are involved in and act in the various developmental periods of different root forms of various plant categories[10−13]. MADS-box TFs are a class of proteins with a MADS-box domain, which is important in regulating plant growth, development and stress responses[14]. Current research on MADS-box transcription factors primarily focuses on their regulatory roles in plant organ developmental programs, particularly floral organogenesis and fruit maturation processes. However, emerging evidence reveals their constitutive expression in understudied compartments including endosperm, pollen grains, guard cells, trichomes, and root vascular tissues, suggesting phylogenetically conserved yet functionally specialized roles in nutrient storage, cellular differentiation, and stress-responsive pathways within these peripheral meristematic domains[15−17].
In recent years, in addition to regulating plant flowers, fruit, seeds and leaf development, an increasing number of MADS-box genes have been confirmed to be involved in the process of root development. Previous studies have shown that the MADS-box transcription factor gene AtANR1, initially identified as the pioneer member of this gene family, orchestrates lateral root elongation in response to heterogeneous nitrate availability through auxin-mediated signaling pathways[18]. The MADS-box TF XAL1/AGL12 has been identified as indispensable for normal root development and growth through its positive regulation of cell cycle-related components[19]. XAL2/AGL14 plays a crucial role in the construction of solid root morphology by regulating auxin polar transport[20], while AGL21 promotes lateral root initiation and growth in Arabidopsis by increasing local auxin biosynthesis[21]. The soybean MADS-box transcription factor GmNMHC5 functions to promote lateral root development through a sucrose-dependent regulatory mechanism[22]. Concurrently, family members of MADS transcription factors (TFs) have been demonstrated to mediate nitrogen signaling-regulated plant growth and development. In rice, the ANR1-like gene OsMADS25 plays a positive regulatory role in the development of primary root (PR) and lateral root (LR) by promoting nitrate accumulation in rice[23]. In barley, the excessive nitrogen-responsive transcription factor HvMADS27 governs root system architecture by modulating abscisic acid (ABA) levels[24]. The enhanced and expanded expression of maize Zmm28 improves photosynthesis and NUE in the crop[25], while truncated MADS TF ZmTMM1 participates in nitrate foraging in maize roots[26]. However, the mechanisms through which MADS-box genes regulate nitrogen uptake and root growth in response to low nitrogen signals remain unclear.
Peach is a perennial fruit tree, and root growth and development is particularly important for perennial trees. In a previous study[27], we identified a peach rootstock 'Shannong-1'(S) which exhibits high NUE. Under controlled growth conditions, Maotao (M)-grafted plants exhibited significantly reduced biomass accumulation compared to 'Shannong-1' rootstock counterparts. A marked improvement in nitrogen utilization efficiency was observed in 'Shannong-1'-grafted specimens. To explore the intrinsic mechanism of high adaptability of S under low nitrogen stress, four cDNA libraries were constructed using 0.1 mM KNO3 and 0.1 mM KCl for 0.5 h. Transcriptomic analysis revealed that genes in multiple pathways within S roots were significantly changed and enriched after low nitrogen treatment, including the nitrogen metabolism pathways. This study identified the low-nitrogen-responsive peach MADS-box transcription factor PpAGL24, elucidated its regulatory mechanisms in root system development and NUE and provides a theoretical foundation for reducing nitrogen input and enhancing NUE in peach orchards.
-
Seeds of Arabidopsis thaliana were surface-sterilized sequentially in 70% (v/v) ethanol (45 s) and 30% (v/v) sodium hypochlorite (3 min), followed by five rinses with sterile distilled water. After stratification at 4 °C for 48 h in darkness, seeds were germinated on Murashige and Skoog (MS) agar plates (0.7% w/v) under controlled conditions (22 °C, 16-h photoperiod). To assess nitrogen responses, plants were subjected to three regimes: Low Nitrogen (LN, 2.94 mM NO3−), Normal Nitrogen (NN, 29.4 mM NO3−), and High Nitrogen (HN, 117.6 mM NO3−).
Peach seeds of roughly the same size were selected and germinated in moist gauze. The germinated seeds were planted on seedling trays filled with quartz sand. Seedlings were grown in plant growth chambers with a photoperiod of 16 h-light (22 °C) and 8 h-dark (18 °C), with a photoperiod of about 200 μmol·m−2·s−1 photonic density and 60% relative humidity, watered only without adding nutrients.
GUS staining
-
The PpAGL24 promoter sequence of proPpAGL24::GUS plasmid, the correct sequencing of vector into Agrobacterium GV3101, Agrobacterium containing proPpAGL24::GUS and Agrobacterium containing empty vector was injected into 4-week old tobacco leaves, 24 h, 16 h/8 h light/dark 28 h, avoid pulse 1 cm leaf punch, 28 °C staining for 24 h, 70% ethanol to the control group green all fade.
Construction of transgenic materials
-
The full-length CDS of PpAGL24 was cloned into a 35s-driven pCAMBIA1300 vector, and the correctly sequenced overexpression vector was transformed into Agrobacterium K599 and transfected into peach trees. When the peach seedlings had six to seven true leaves, the entire root was cut and transfected through the wound. The transformed peach seedlings were put back into quartz sand for continued cultivation and root development.
The AGL 24 mutant obtained the T-DNA insertion line SALK_026551 from the Arabidopsis Biological Resource Center (ABRC), identified the homozygous mutant strain by RT-PCR using gene-specific primers AGL24 LP, AGL24 RP and universal primer BP. Related primers were synthesized for overexpressed Arabidopsis thaliana (OE) in Supplementary Table S1. PpAGL24 Overexpressed Arabidopsis thaliana (OE) was provided by oebiotech Company.
Phenotypic identification of the root system morphology
-
Root morphology was examined on MS medium solidified with 0.7% (w/v) agar. Seeds were grown horizontally on MS medium and 3 d after germination, seedlings with similar growth status were selected and transferred to 13 × 13 cm2 MS medium, LN medium, NN medium, and HN medium for vertical growth. From the day of transplanting, the number of visible lateral roots and the length of primary roots were photographed after 6 d, and the root length of digital images of the plants was measured manually using ImageJ software (NIH).
Root phenotypic analysis was performed on peach seedlings at 25 d post-infection (PpAGL24-GFP overexpression lines). GFP fluorescence in root tissues was visualized using a handheld fluorescence lamp (LUYOR-3415RG, LUYOR, USA). Roots exhibiting distinct green fluorescence signals were identified as GFP-positive transgenic roots, and entire root systems meeting this criterion were collected as uniform samples for phenotypic characterization and physiological parameter measurements. Transformation efficiency was calculated as the ratio of plants with GFP-expressing roots to the total number of infected plants, with each transgenic line infected using a minimum of 60 peach seedlings. GFP-positive root systems were subsequently subjected to scanning and quantitative analysis using a root imaging system[28].
Plant genomic RNA extraction
-
The expression of PpAGL24 in the transgenic roots was determined by RT-qPCR 24 d after peach seedling transformation. Transcriptome sequencing was performed on roots derived from control and successfully overexpressed transgenic lines. Total RNA was extracted using the TRIzol reagent (Invitrogen). Three biological replicates were performed for each sample. RNA purity and quantification were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) and stored in an ultra-cold refrigerator for gene expression analysis.
Quantitative PCR analysis of reverse transcription
-
Total RNA from plant root samples was extracted with TRIzol (Invitrogen) and reverse transcribed as cDNA using PrimeScript RT kit (Takara, Japan). PpActin and AtActin were used as the reference genes for peach and Arabidopsis thaliana, respectively. Gene-specific primers were designed using NCBI. The RT-qPCR was performed using SYBR Premix Ex Taq (Takara, Japan) with three biological and three technical replicates on a QuantStudio 3 real-time PCR instrument (Thermo, USA). All the reagents were used according to the manufacturer's instructions. Relative expression of genes was calculated using 2−ΔΔCᴛ to analyze the resulting data. Relevant primers are listed in Supplementary Table S1.
SPAD value measurement
-
Leaf relative chlorophyll content was measured non-destructively using a SPAD-502 Plus portable chlorophyll meter (Konica Minolta, Japan). For each plant line, nine plants were randomly selected, and fully expanded functional leaves from the rosette leaf disc were chosen for measurement, ensuring the absence of visible disease/pest damage or mechanical injury.
Dual-luciferase assay
-
The coding sequence of PpAGL24 was reconstituted into 35S, the driven pGreenII 62-SK vector, and the promoter sequences of PpNR, PpNiR1, PpNPF1.2, and PpNRT2.1 were individually cloned as reporter genes into the pGreenII0800-LUC vectors. The correctly sequenced vectors were transformed into Agrobacterium GV3101. Agrobacterium containing pGreenII 62-SK-PpAGL24 and reporter gene were injected into leaves of Nicotiana as the experimental group, and multiple control groups were injected into different regions of the same leaf. Two days later, the back side of the leaves was sprayed with a 1 mM D-fluorescein solution of sodium salt. Luciferase signals were collected and analyzed for luciferase signals using the IVIS LuminaII In vivo Imaging Facility (Xenogen, USA). Luciferase activities were quantified using a dual-luciferase reporter assay kit. Firefly luciferase (LUC) activity values were normalized to Renilla luciferase (REN) internal control signals (expressed as LUC/REN ratio) and subsequently standardized against the empty vector control group. Related primers are listed in Supplementary Table S1.
Yeast one-hybrid assay
-
The coding sequence of PpAGL24 was cloned into the pGADT7 vector (AD-PpAGL24) and the promoters of PpNR, PpNiR1, PpNPF1.2, and PpNRT2.1 were cloned into the pHISII vector. Either pGADT7 or AD-PpAGL24 was then transferred into the yeast Y187. The pGADT7 empty vector together with pHISII-PpNRpro, PpNiR1pro, PpNPF1.2pro, PpNcd-2pro, and PpNRT2.1pro or pHIS as a control. Yeast cultures were grown on a selective medium (-Trp/-Leu/-His) at 30 °C for 72 h. Colonies demonstrating standard growth morphology with consistent size were resuspended to achieve a standardized cell density (OD600 = 0.2/0.0002). Aliquots of 7 μL from the resuspended cultures were inoculated onto a selective medium (-Trp/-Leu/-His) supplemented with 80 mM 3-amino-1,2,4-triazole (3-AT) to inhibit background autoactivation. The pGADT7 empty vector served as the negative control. Post-incubation colony development was systematically evaluated to determine interaction specificity. Relevant primers are listed in Supplementary Table S1.
Statistical analysis
-
All experiments were performed with at least three independent biological replicates unless otherwise stated. Data are presented as mean ± SD. Statistical significance between experimental groups was determined using Duncan's multiple range test and Student's t-test (p < 0.05), with significant differences indicated by distinct lowercase letters following the results of multiple comparison analyses.
-
To further identify the key factors involved in high NUE in S, we compared gene expression between S and Maotao (M) after nitrogen treatment. The transcription factor families with the most differentially expressed genes (DEGs) were MIKC-MADS (MIKC-type MADS-domain proteins) TFs, Dof (DNA binding with one figure) TFs, ERF (Ethylene responsive factor) TFs, and MYB (V-myb avian myeloblastosis viral oncogene homolog) TFs (Fig. 1a). MADS-box transcription factors have been shown to play important roles in plant root development and nitrogen response; however, their family members have not been extensively studied in peach, particularly in subterranean tissues. Therefore, we focused on the MADS family, and through RT-qPCR validation of the transcriptome data, we found that the expression of PpAGL24 was up-regulated at higher folds in S under KNO3 treatment compared with that of peach (M) rootstocks (Fig. 1b), and then we selected PpAGL24, which had the highest folds, as the focus of our study. To investigate the nitrogen-dependent response of PpAGL24, we performed RT-qPCR and GUS staining assays using the proPpAGL24::GUS reporter line under varying nitrogen regimes (Fig. 1c, d). The results showed that PpAGL24 promoter was able to respond to low nitrogen signal, and the transcriptional activity of the PpAGL24 promoter was significantly enhanced under low nitrogen conditions, but with no significant alteration in transcriptional activity at medium and high nitrogen levels. This suggests that PpAGL24 can respond to low N signals and may play a role in the S high NUE.
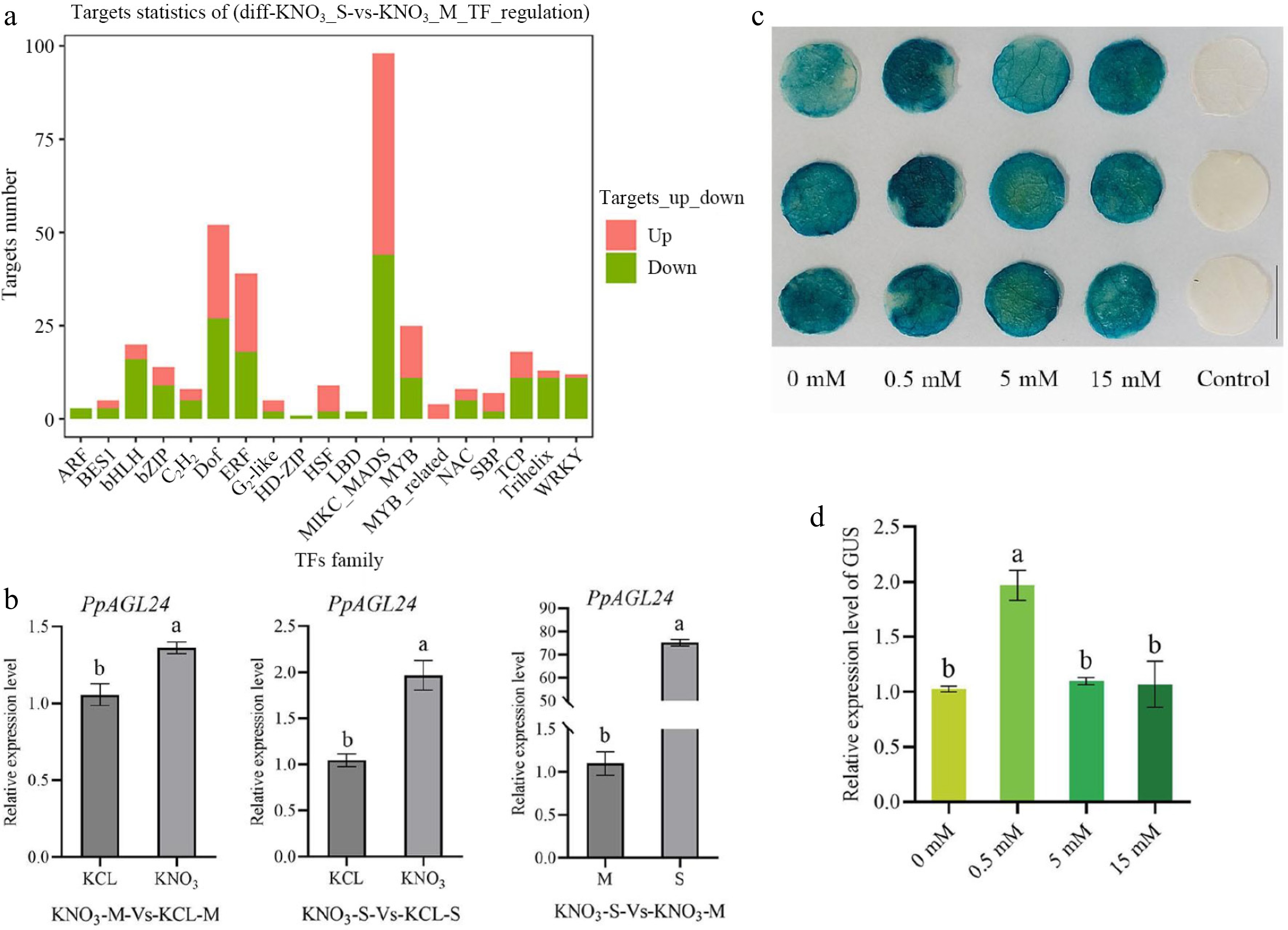
Figure 1.
Transcription factor analysis of differentially expressed transcription factors in 'Shannon-1' (S) and Maotao (M) rootstocks after low nitrogen treatment. (a) TF family with significant DEGs in roots of S and M after 0.1 mM KNO3 treatment. (b) Differential relative expression levels of PpAGL24 in S and M tissues after treatment with 0.1 mM KNO3. Relative transcript levels were normalized to KCl-treated controls (for S) or M baseline (for M), respectively, and expressed as mean ± SD (n = 3). Statistical significance (p < 0.05, Student's t-test) is denoted by distinct lowercase letters above bars. (c) Construct proPpAGL24::GUS reporter infected four-week-old tobacco, performed 0, 0.5, 5, and 15 mM KNO3 100 mL root solution at 0, 24, and 48 h after infection, took 1 cm leaf discs for GUS staining 52 h after infection, and decolorized at 28 °C overnight. The Control group used tobacco leaves injected with a bacterial solution containing empty vectors. (d) Relative expression levels of the GUS gene in tobacco leaf discs were analyzed by RT-qPCR, with transcript levels normalized to the 0 mM KNO3− treated control (GUS). Data are expressed as mean ± SD (n = 3). Distinct lowercase letters indicate statistically significant differences (p < 0.05) determined by one-way ANOVA followed by Duncan's multiple range test.
Analysis of the cis-acting elements on the PpAGL24 promoter
-
We conducted a systematic cis-acting element analysis of the PpAGL24 promoter using the PLANT CARE platform. The results revealed that the upstream promoter region of PpAGL24 harbors a multitude of cis-acting elements, primarily including hormone-responsive elements, light-responsive motifs, group and development-related, and stress-related regulatory sites (Supplementary Fig. S1). Among the hormone response elements are auxin response element, gibberellin response element, cis-acting elements involved in gibberellin response, and homeopathic elements involved in abscisic acid response. This suggests that PpAGL24 may regulate plant growth and development by integrating polyhormonal signalling and environmental factors.
Overexpression of PpAGL24 regulates root growth in Arabidopsis thaliana and increases lateral root number
-
As auxin plays an important role in root growth and development[29,30], therefore, we initially used Arabidopsis as a model system to investigate the possible role of PpAGL24 in root growth and NUE. We obtained the 35S::PpAGL24 transgenic Arabidopsis plants and pure and mutants of the T-DNA insertion: SALK_026551 (agl24). To assess the impact of PpAGL24 on NUE and root system development, 35S::PpAGL24 overexpression lines (OE1, OE2), AGL24 mutants (agl42-1, agl24-2), and Col-0 wild-type plants were vertically grown on Murashige and Skoog (MS) medium. Phenotypic and metabolic responses were analyzed under controlled nitrogen regimes to elucidate genotype-specific regulatory mechanisms. Briefly, Arabidopsis plants with different genotypes were seeded on MS medium, cultured normally after 2 d of 4 °C vernalization (16 h light/8 h dark), transplanted to Normal Nitrogen, Low Nitrogen, and High Nitrogen (NN, LN, and HN) media after 4 d, and root phenotypes were observed for 7 d of vertical growth. Regardless of nitrogen supply, Arabidopsis overexpressing PpAGL24 showed better root development than wild-type plants, mainly through an increased number of lateral roots. The mutants also showed a decrease in the number of lateral roots but no significant difference in primary root length between the lines (Fig. 2a−e). The death of mutants on HN medium may be due to weakened expression of related genes. In addition, after overexpression of PpAGL24, the shoots of the overexpression lines showed greater rosette leaf area and higher SPAD values compared with wild type (Fig. 2f−i), as well as a significant increase in shoot dry weight and fresh weight (Supplementary Fig. S2), which is also a manifestation of root optimization and improved NUE.
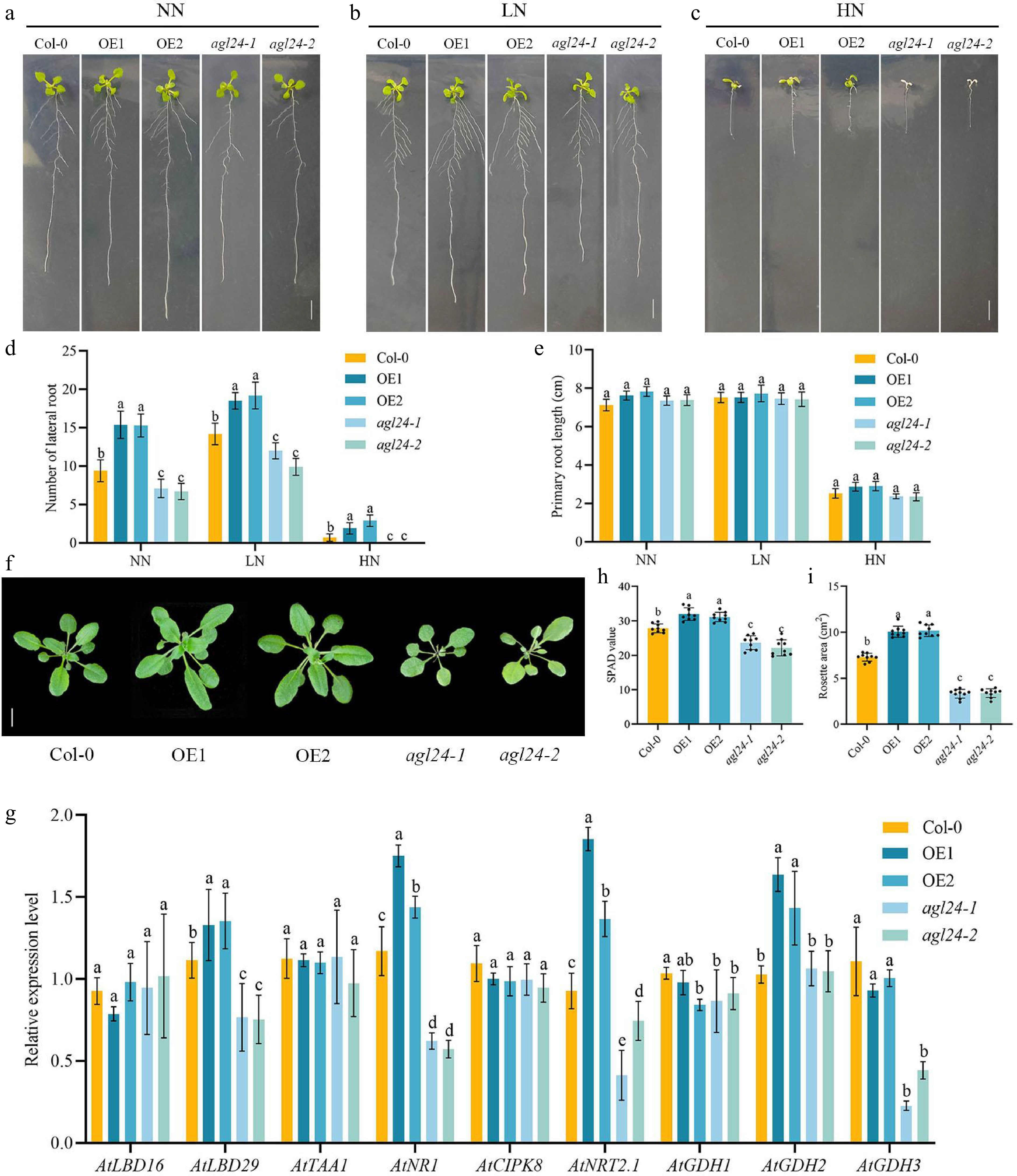
Figure 2.
Analysis of phenotypic characteristics and nitrogen metabolism-related gene expression in Arabidopsis heterologously overexpressing PpAGL24. (a)−(c) Arabidopsis seedlings from overexpressing lines (OE1 and OE2), wild-type Col-0, and AGL24 mutants (agl24-1 and agl24-2) 3 d after germination in MS medium were transplanted to root phenotypes for 7 d in NN, LN, and HN. (d), (e) Number of lateral root length and primary root in Arabidopsis under NN, LN, and HN conditions. (f) Col-0, OE1, OE2, agl24-1, and agl24-2 20-day-old Arabidopsis thaliana shoots. (g) Relative expression levels of genes involved in root development and nitrogen uptake utilization in Col-0, OE1, OE2, agl24-2, and agl24-2 Arabidopsis under NN conditions. (h), (i) SPAD values and rosette leaf area of 20-day-old Arabidopsis. Data represents means ± SD from three independent biological replicates. Significant differences were determined using Duncan's multiple range tests (p < 0.05).
To further validate the functional role of PpAGL24 in NUE and root system development, we analyzed the effects of PpAGL24 overexpression on the expression of NUE and root development-associated genes in Arabidopsis. Notably, the expression of AtLBD29 (Lateral organ boundaries domain 29), AtNR1 (Nitrate oxidoreductase), AtGDH2 (Glutamate Dehydrogenase, GLDH2), and AtNRT2.1 (Nitrate transporter 2.1) was significantly induced in overexpression lines, while mutation of AtAGL24 exerted a repressive effect on their transcriptional activity (Fig. 2g). These results suggest that overexpression of PpAGL24 likely modulates Arabidopsis root system development and NUE by regulating the expression of related genes.
Overexpression of PpAGL24 promotes peach root development, nitrogen uptake, and utilisation
-
The PpAGL24 gene was cloned from the root system of the peach rootstock 'Shannong-1' (S). Overexpression of this gene in peach roots was achieved through an Agrobacterium rhizogenes strain K599-mediated root transformation system. RT-qPCR analysis demonstrated a 9.729-fold increase in PpAGL24 expression levels in the overexpressing roots (Pro35S:PpAGL24-GFP) compared to the control group (Fig. 3a). Transformation efficiency analysis showed that the transformation efficiencies of regenerated root systems in the Control and Pro35S:PpAGL24-GFP groups were 33.12% and 46.63%, respectively (Supplementary Fig. S4), and the higher transformation efficiencies increased the reliability of the present experimental system in gene function studies. Upon analyzing the regenerated root systems (Fig. 3b), PpAGL24 exhibited a 66.67% and 95.59% increase in root length and root tips respectively, while root volume and dry weight increased by 59.09%, and 78.36% respectively. Additionally, the total nitrogen content increased by 52.82% and nitrate by 11.22% after overexpression of PpAGL24 in the regenerated roots(Fig. 3c−i). These results functionally demonstrate that PpAGL24 positively regulates effect on peach root lineage development and can influence root growth and promote root nitrogen uptake and utilization.
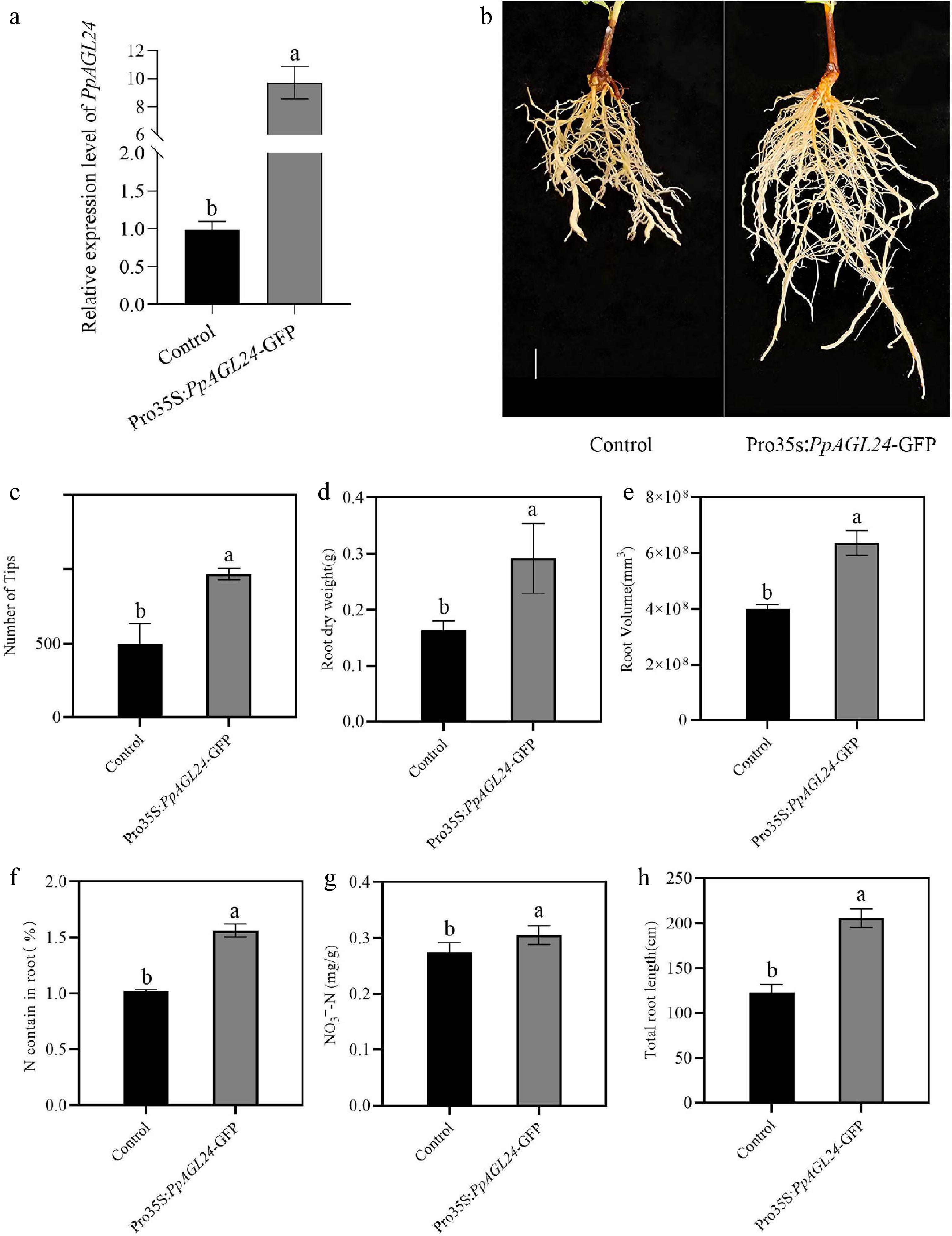
Figure 3.
Overexpression of PpAGL24 promotes peach root development and nitrogen uptake. Pro35s:PpAGL24-GFP represents overexpressed peach roots of PpAGL24, Control represents control transgenic peach roots containing the empty vector. (a) RT-qPCR examination of PpAGL24 transcript levels within control and transgenic root systems. Relative transcript levels were normalised to Control, expressed as means ± SD, n = 3. (b) Phenotypic characterization of regenerated peach roots following Agrobacterium rhizogenes strain K599-mediated transformation. Scale bar = 1 cm. (c)−(i) Number of tips, root dry weight, root volume, N contain in root, NO3−-N contain, total root length. Statistical significance (p < 0.05, Student's t-test) is denoted by distinct lowercase letters above bars.
Transcriptomic analysis of peach root after overexpression of PpAGL24
-
To elucidate the molecular mechanisms underlying PpAGL24-mediated root development regulation and NUE enhancement in the peach rootstock 'Shannong-1', we constructed two cDNA libraries using the p1300-Empty and p1300-PpAGL24 constructs through high-throughput sequencing. After the removal of the adapter and low-quality sequences, we obtained clean reads Q30 > for 47.35 M (AGL) and 47.61 M (CK), 96.24%, and 96.20%, respectively. Principal component analysis (PCA) showed good reproducibility between the three biological replicates (Fig. 4a). Volcano plots show the relationship between the significance of the p-values and the fold change of all the differentially expressed broad universal genes (DEGs). The heat map of DEGs visually depicts the expression between CK and roots overexpressing PpAGL24, where the red color indicates the high expression levels, blue color indicates the low expression levels. Differential expression analysis of genes showed that, compared to CK, 325 genes were significantly differentially expressed after overexpressing PpAGL24. The red scatter represents 200 upregulated genes (AGL relative to CK), blue scatter points represent 125 downregulated genes (Fig. 4b), subsequent pathway enrichment and functional analysis of these genes were performed.
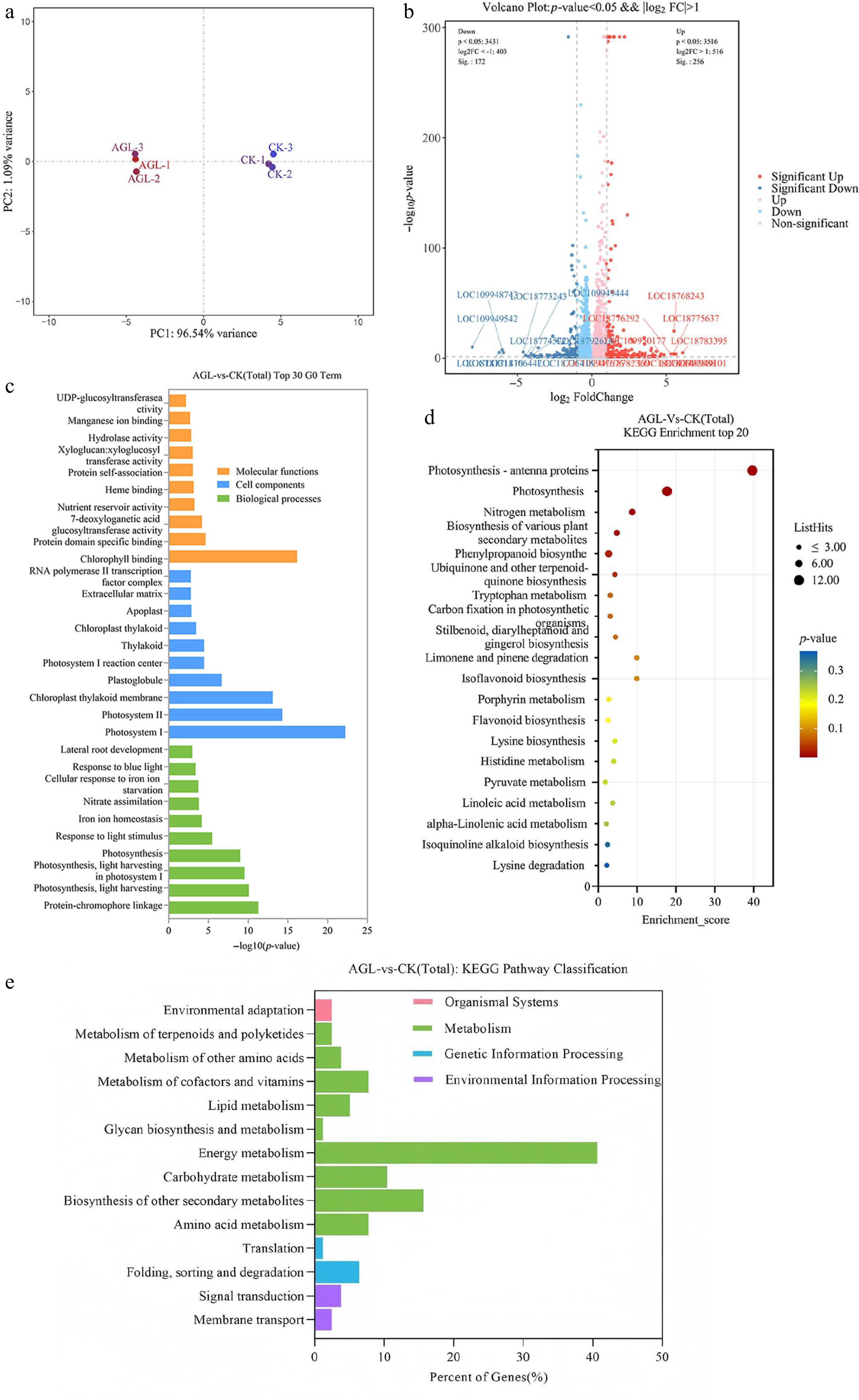
Figure 4.
Comparison of the two transcriptomic libraries. (a) Principal component analysis of the two transcriptomes, AGL-1, AGL-2, and AGL-3: Pro35s: Three biological replicates of peach root overexpressing the PpAGL24-GFP. CK-1, CK-2, and CK-3: three biological replicates of control transgenic peach containing empty vector. (b) Genes differentially expressed in AGL, red scatter are genes upregulated in AGL relative to CK, and blue scatter are downregulated genes. (c) The GO classification of the differentially expressed genes. (d), (e) KEGG pathway enrichment for the differentially expressed genes.
These DEGs were annotated with GO functions, screening the GO entries corresponding to PopHits ≥ 5 in the three categories (cell components, molecular functions, and biological processes), sorting the top −log10p-value for each entry. In the cell component category, DEGs were relatively high in the 'photosystem I', 'photosystem', and 'chloroplast thylakoid membrane' types. Among the molecular function categories, 'chlorophyll binding' and 'nutrient storage activity' were relatively high. Meanwhile, the GO terms of 'photosynthesis light capture', 'nitrate assimilation', and 'lateral root development' in the biological process category were abundant (Fig. 4c). Notably, the expression of PpAGL24 within the peach root lineage caused significant changes in gene transcripts related to 'nitrate assimilation' and 'lateral root development'.
In the Pathway enrichment analysis of DEGs, the genes were mainly enriched in the four categories: environmental information processing, genetic information processing, metabolism, and organic system. Among these, DEGs were the most enriched in metabolism, accounting for 84.88%, while DEGs enriched in the energy metabolism pathway accounted for 42.27%. Figure 4 shows the 20 pathways with the largest number of DEGs in the metabolic classification, mainly including Photosynthesis-antenna proteins, photosynthesis, nitrogen metabolism, and the synthesis of various secondary metabolites in plants (Fig. 4d, e).
At the same time, the qRT-PCR results showed enrichment in the nitrogen metabolism pathway and root development pathway-related gene expression in AGL and CK with significant difference (Supplementary Fig. S3). This result is consistent with the transcriptome data, which suggests that the expression of PpAGL24 may affect energy metabolism and root development within the peach root system, causing genes on the nitrogen metabolism pathway to function, which in turn has an impact on plant nitrogen utilisation and root development.
PpAGL24 promotes PpNiR1 expression by binding to its promoter
-
To further investigate the molecular mechanism by which PpAGL24 positively regulates peach root development and nitrogen uptake/utilization, we selected five nitrogen metabolism-related genes positively regulated by PpAGL24 through GO functional annotation and KEGG pathway enrichment analysis of differentially expressed genes (DEGs) in the transcriptome. Dual-luciferase assays revealed that PpAGL24 significantly activated the transcriptional activity of the PpNiR1 promoter (Fig. 5a, b). Subsequently, yeast one-hybrid experiments demonstrated the specific binding capacity of PpAGL24 to the PpNiR1 promoter region (Fig. 5c). Collectively, these findings establish that PpAGL24 transcriptionally activates PpNiR1 expression through promoter interaction.
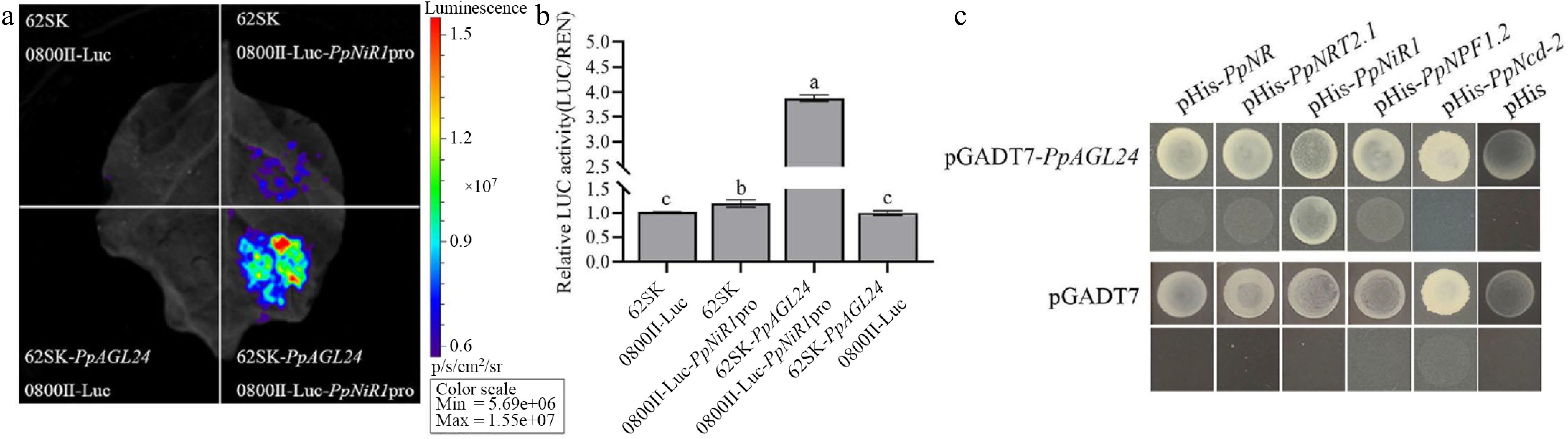
Figure 5.
PpAGL24 is able to activate PpNiR1 transcription. (a) Luciferase imaging analysis. (b) Relative luciferase activity was normalised to LUC/REN for 0800II-Luc empty vector + 62SK empty vector by measuring luciferase activity, expressed as mean ± SD, n = 3. (c) Yeast one-hybrid experiments showed that PpAGL24 was able to bind the promoter of PpNiR1. Different lowercase letters indicate significant differences at p < 0.05 by one-way ANOVA and Duncan (D) multiple comparisons test.
-
Nitrogen (N), an essential macronutrient for plant growth, predominantly exists in soil as nitrate and ammonium[31]. Nitrogen plays critical roles in multiple physiological processes during plant growth and development, including the biosynthesis of proteins and nucleic acids, and serves as a key limiting factor for plant growth and agricultural productivity[32]. Roots are an important organ for plants to acquire soil nutrients[33]. For perennial fruit trees, robust and well-developed root systems play a crucial role in nutrient uptake[34]. Plant root systems are equipped with sophisticated nutrient uptake mechanisms that dynamically respond to soil nutrient signals and orchestrate associated physiological processes. Nitrogen acquisition in plants is predominantly mediated through root-mediated absorption from the soil environment. Therefore, elucidating the regulatory mechanisms governing root system development and nitrogen utilization dynamics is imperative for optimizing nitrogen use efficiency in plants, which holds strategic significance for sustainable agriculture. In this study, PpAGL24 was identified as a MADS-box transcription factor (TF) from the high nitrogen use efficiency (NUE) peach rootstock 'Shannong-1', exhibiting regulatory potential in nitrogen metabolism. Functional characterization and transcriptional activation assays revealed that PpAGL24 responds to low nitrogen availability, positively modulates root system architecture, enhances NUE, and ultimately coordinates plant developmental processes.
The peach system primarily uses nitrate nitrogen in the soil. Previous studies showed that 'Shannong-1' (S) is a peach stock with high NUE. To explore the reasons of the high NUE, the roots of the two stocks were treated with KCl and KNO3 followed by transcriptome sequencing[27]. Among the most abundant DEGs in the two stocks after nitrogen treatment were the MADS-box TF family. Previous studies have demonstrated that MADS-box transcription factors regulate root system architecture remodeling, optimizing nitrogen use efficiency (NUE) in plants. In Arabidopsis, AGL17/AGL21, and ANR1 belong to the AGL 17 clade and are preferentially expressed in roots where they respond to nitrogen signals involved in lateral root formation and development[18,21]. Tomato SlMBP9 affects lateral root formation by modulating expression of auxin content in the root region[35]. This suggests that MADS-box TF play a role in root development. In this study, we observed that heterologous or homologous overexpression of PpAGL24 positively regulated root development. Homologous overexpression of PpAGL24 in peach roots using Agrobacterium rhizogenes K599 significantly increased root biomass and improved root system architecture. Transcriptome analysis combined with GO functional annotation showed that PpAGL24 negatively regulated the expression of lateral root development genes: PpWRKY75/LOC18777012 (a direct homologue of the Arabidopsis root growth inhibitor AtWRKY75/AT5G13080[36]), PpIP5P12/LOC18770023, and PpTAA1/LOC18792031. PpAGL24 may contribute to the development of the root system by alleviating the inhibitory pathway. In the model plant Arabidopsis thaliana, heterologous expression of the PpAGL24 gene resulted in a significant increase in root biomass and optimisation of root conformation under different nitrogen supply conditions. The transcript level of AtLBD29/AT3G58190, a regulatory factor involved in lateral root development[37], was altered by the ectopic expression of PpAGL24. This suggests that PpAGL24 can still fulfill its regulatory function in heterologous systems. This finding, together with previous findings in peach, reveals the conserved biological function of the PpAGL24 gene across species and its molecular mechanism of root plasticity improvement through the regulation of key repressors, which positions PpAGL24 as a key positive regulator that promotes root growth and development by coordinating the repression of developmental constraints.
Transcription factors regulate the expression of downstream genes by identifying and binding to specific motifs. In rice, OsMADS57 regulates nitrate transport from the subbasement to the subshoot by binding to the CArG motif on the OsNRT2.3a promoter[38]. In our study, overexpression of PpAGL24 enhanced nitrogen utilization in roots, and the transcriptome of PpAGL24 transgenic roots showed enrichment of the differential genes in the 'nitrate assimilation' pathway, which was functionally annotated as 'nitrogen metabolism'. RT-qPCR also showed significant changes in the expression of nitrogen transport proteins PpNRT2.1 and PpNPF1.2, as well as nitrogen assimilation-related genes such as PpNR and PpNiR1. This suggests that PpAGL24 may further affect nitrogen uptake and utilization by influencing the expression of nitrogen metabolism-related genes. Subsequent yeast one-hybrid (Y1H) and dual-luciferase reporter assays demonstrated that PpAGL24 binds to the PpNiR1 promoter and activates its transcriptional activity. Ectopic expression of PpAGL24 in Arabidopsis caused significant changes in expression of AtNR1, AtNRT2.1, etc. This all suggests that PpAGL24 can influence NUE in plants.
In the transcriptome analysis of PpAGL24 overexpressing peach roots, we found that differentially expressed genes (DEGs) showed significant enrichment in photosynthesis-related pathways. Given that nitrogen is a key element for chlorophyll biosynthesis, there may be a synergistic regulatory relationship between enhanced photosynthetic capacity and enhanced nitrogen utilisation efficiency, which provides new evidence that PpAGL24 optimises nitrogen utilisation efficiency through system-level metabolism. Of particular interest is the up-regulated expression of genes encoding key components of the photosynthetic electron transport chain (e.g., PpPSAH/LOC18784475, PpPSBT/LOC18780376)[39] and the CAB family of chlorophyll-binding proteins[40], suggesting that this regulatory process may establish a balance of carbon and nitrogen metabolism by enhancing the efficiency of the light-dark coupling reaction. However, how the local overexpression of PpAGL24 in the root system remotely regulates the molecular module of leaf photosynthesis, and its long-distance signalling mechanism (e.g. hormone signalling or small RNA-mediated systemic regulation) remains to be further resolved.
In conclusion, our study demonstrates that PpAGL24 enhances root system development in peach, particularly through promoting lateral root formation, and transcriptionally modulates the expression of PpNiR1 encoding a nitrite transporter protein, ultimately improving nitrogen absorption and assimilation efficiency. These findings provide a theoretical framework for optimizing nitrogen management in peach orchards through reduced fertilizer input while maintaining productivity and offer novel insights into the functional diversification of MADS-box transcription factors in peach.
This work was supported by the China Agriculture Research System-Peach industry (CARS-30-2-02), and the National Natural Science Foundation of China (32272651).
-
The authors confirm contribution to the paper as follows: study conception and design: Peng F, Xiao Y, Yu C; data collection: Cai H, Liang J, Yu H, Huang F, Wang H; analysis and interpretation of results: Fan S, Wang Z, Guo J, Luo J, Chen Q, Gao H, Gao Y, Wu X; draft manuscript preparation: Xiao Y, Peng F, Yu C, Cai H, Ngomuo RS. All authors reviewed the results and approved the final version of the manuscript.
-
The data that support the findings of this study are available in the NCBI repository.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 Related PCR primer sequences.
- Supplementary Fig. S1 Analysis of cis-acting elements of the PpAGL24 promoter.
- Supplementary Fig. S2 Aboveground fresh and dry weights of Arabidopsis thaliana in Col-0, OE1, OE2, agl24-1, and agl24-2 genotypes.
- Supplementary Fig. S3 RT-qPCR validation of relative expression levels of related genes within peach roots after homologous overexpression of PpAGL24.
- Supplementary Fig. S4 Transformation efficiency of K599-infested peach root systems. Statistical significance (p < 0.05, Student's t-test) is denoted by distinct lowercase letters ab ove bars.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Yu C, Cai H, Liang J, Wu X, Yu H, et al. 2025. The regulatory mechanism of the low-nitrogen-responsive MADS-box transcription factor PpAGL24 in peach root system development and nitrogen utilization. Fruit Research 5: e020 doi: 10.48130/frures-0025-0015
The regulatory mechanism of the low-nitrogen-responsive MADS-box transcription factor PpAGL24 in peach root system development and nitrogen utilization
- Received: 29 January 2025
- Revised: 06 May 2025
- Accepted: 07 May 2025
- Published online: 19 May 2025
Abstract: Nitrogen critically regulates peach yield formation and fruit quality development through multifaceted physiological mechanisms. In peach production, the problems of large nitrogen input and low Nitrogen Use Efficiency (NUE) are more prominent. Therefore, mining key genes for efficient nitrogen utilization in peach is crucial for improving NUE and reducing nitrogen use. Integrated transcriptomic profiling of peach rootstock 'Shannong-1' under low-nitrogen (0.1 mM) treatment identified the MADS-box transcription factor PpAGL24. Functional characterization revealed that PpAGL24 modulates root system architecture and coordinates nitrogen uptake-assimilation pathways, suggesting its pivotal role in enhancing NUE. PpAGL24 expressed in peach roots and overexpressed in Arabidopsis thaliana both resulted in an increased number of lateral roots and higher nitrogen content. Transcriptome sequencing of PpAGL24-overexpressing peach roots revealed its putative regulatory network governing 'lateral root development', 'nitrate assimilation', and 'nitrogen metabolism', suggesting its hierarchical coordination in root developmental plasticity and nitrogen remodeling. Dual-luciferase and yeast one-hybrid assays showed that PpAGL24 can activate the transcription of the nitrite reductase-encoding gene PpNiR1. This study reveals a new mechanism by which the MADS-box transcription factor PpAGL24 regulates root growth and nitrogen assimilation in peach trees by modulating the PpNiR1 gene, which provides theoretical support for the improvement of NUE and the reduction of nitrogen inputs in peach orchards.
-
Key words:
- Peach /
- PpAGL24 /
- Root growth /
- NUE













