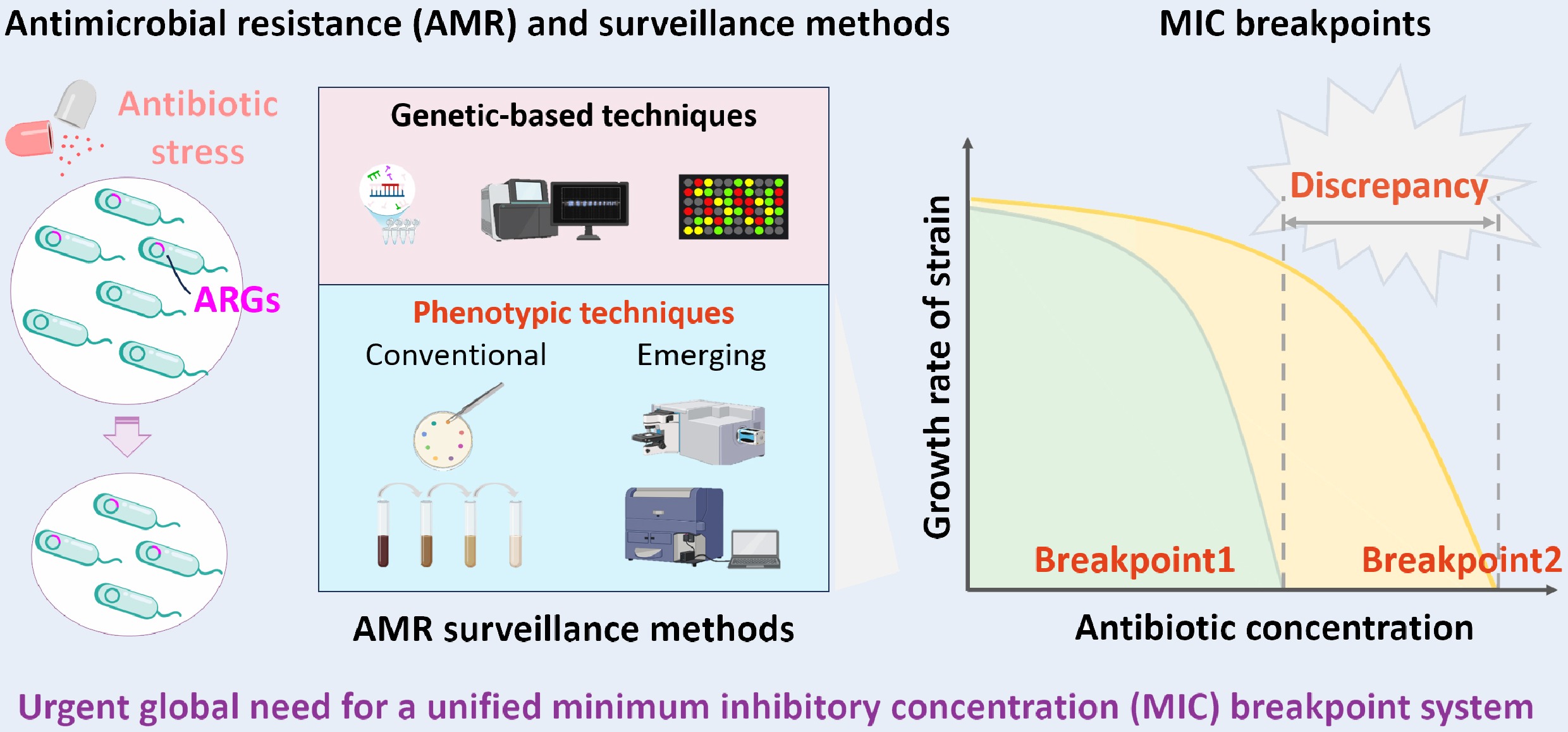
-
Since the discovery of penicillin in 1929, antibiotics have been extensively utilized in medical, agricultural, and livestock production, significantly contributing to global public health and socioeconomic development[1]. Global antibiotic consumption surged by 65% between 2000 and 2015, with projections indicating a further 200% growth by 2030[2]. Due to their environmental persistence and continuous discharge, antibiotics have been ubiquitously detected across various ecosystems, with median concentrations ranging from not detected (ND) to 286 ng/L in global rivers[3], 20.7–43.3 ng/L in freshwater reservoirs of Europe and Asia[4], 0.71‒27.5 ng/L in global groundwater[5], and with individual mean concentrations up to 10.00 ng/g dry weight in agricultural soils based on 2,225 observations from 135 independent studies[5,6]. Residual antibiotics persistently accumulating in ecosystems pose significant threats to aquatic and terrestrial organisms, potentially reducing biodiversity, disrupting biogeochemical cycles, and breaking ecological balances[7]. Furthermore, these compounds bioaccumulate through food chains, ultimately reaching human bodies where they may induce toxicological effects and create long-term health risks[8].
Antibiotic pollution poses a dual threat as both an environmental disruptor, and a toxicological hazard to human health, while also accelerating the emergence and spread of antimicrobial resistance (AMR), and antibiotic resistance genes (ARGs)[9]. A study conducted at three large-scale commercial swine farms in China identified 149 unique ARGs, with the top 63 ARGs showing enrichment ranging from 192-fold (median) to 28,000-fold (maximum), compared to antibiotic-free controls[10]. In addition to antibiotics at environmentally relevant concentrations[11], studies have found that even at sub-inhibitory levels, which are several hundred-fold below the minimal inhibitory concentration (MIC) of susceptible bacteria, can also enrich antibiotic resistant bacteria (ARB)[12−14]. In activated sludge reactors treating antibiotic production wastewater, exposure to multiple antibiotics significantly increased the abundance and proportions of plasmid-mediated ARGs (57.9%)[15]. These processes further promote the selection and proliferation of antibiotic resistance through horizontal gene transfer (HGT) in the environment[16]. A global investigation revealed that high-risk ARGs (Rank I) have increased by 2.0–36.3 times in soils and clinical isolates compared to pre-2010 levels, indicating a rising environmental and public health threat[17]. The ability of clinically significant ARGs to cross habitat boundaries complicates treatment regimens and compromises therapeutic efficacy[18]. In 2019, antibiotic resistant infections caused an estimated 4.95 million fatalities worldwide[19]. Without immediate intervention, AMR-related mortality could escalate to 10 million deaths per year by 2050, with cumulative economic losses exceeding 100 trillion USD[20]. Therefore, the World Health Organization (WHO) has listed antibiotic resistance as one of the top ten global public health threats, calling for strict regulatory and management strategies[7].
Effective environmental regulatory policies for AMR control depend on rapid, accurate, and cost-effective surveillance technologies. Over the past century, primary categories of techniques have been developed to evaluate the dissemination of AMR and their resistance levels, including: (1) genetic-based techniques (such as polymerase chain reaction (PCR), quantitative PCR (qPCR), and metagenomic sequencing[21,22]), and phenotypic detection methods; (2) conventional methods (such as disk diffusion, broth dilution, and E-test[23−29]); and (3) emerging methods (such as Raman spectroscopy and microfluidic technologies[30−33]). The genetic-based techniques can rapidly identify the presence of ARGs. In comparison, phenotypic detection methods provide the MIC through direct observation of bacterial growth inhibition in the presence of a gradient of antibiotic concentrations[34]. The MIC value serves as the gold standard in antibiotic susceptibility testing, providing a critical metric for assessing resistance in both clinical and environmental settings, as well as for guiding the development of new antimicrobial agents[35]. Nevertheless, interpretive discrepancies exist between major standards, such as the Clinical and Laboratory Standards Institute (CLSI), and the European Committee on Antimicrobial Susceptibility Testing (EUCAST), potentially affecting clinical decisions and environmental resistance surveillance[36].
Therefore, the primary objectives of this review are to: (1) systematically examine the fundamental principles, technical advantages, and limitations of the latest AMR surveillance technologies; and (2) analyze the key divergences based on major active standards, paying more attention to the differences between CLSI and EUCAST interpretive criteria. This study strongly advocates for the establishment of a more authoritative and unified framework to provide more precise guidance for global antibiotic resistance monitoring and antimicrobial drug evaluation.
-
Current AMR surveillance approaches are broadly categorized into genetic-based detection methods (identifying resistance genes through molecular methods), conventional phenotypic detection methods (relying on microbial growth inhibition), and emerging phenotypic detection methods (leveraging novel biosensing and imaging strategies). Among these approaches, genetic-based detection methods for AMR surveillance primarily target resistance genes. They offer rapid and highly sensitive approaches, enabling faster diagnosis and early intervention. However, they are unable to directly assess the inhibitory effect of antibiotics on bacterial growth. Accordingly, the value of MIC can only be directly detected through phenotypic detection methods, including dilution methods, E-test, automated systems, microfluidic platforms, electrochemical assays, and fluorescence-based detection[34]. The most widely used MIC determination method is the broth microdilution method, which measures the lowest concentration of an antibiotic required to inhibit bacterial growth. This approach is highly standardized and applicable to most bacterial tests, but it consumes a large number of reagents, and is susceptible to false positives or cross-contamination[37,38]. With the advancement of technology, AMR surveillance methods that primarily rely on optical, molecular biological, and metabolic activity techniques can achieve antibiotic susceptibility testing in a shorter period. These technologies can achieve far greater speed and precision than traditional culture-based methods[31]. However, their high equipment costs and complex operational procedures limit widespread adoption in routine laboratory settings. The following subsection systematically examines these methodologies, highlighting their respective principles, advantages, and limitations in clinical and research applications (Fig. 1, Table 1).
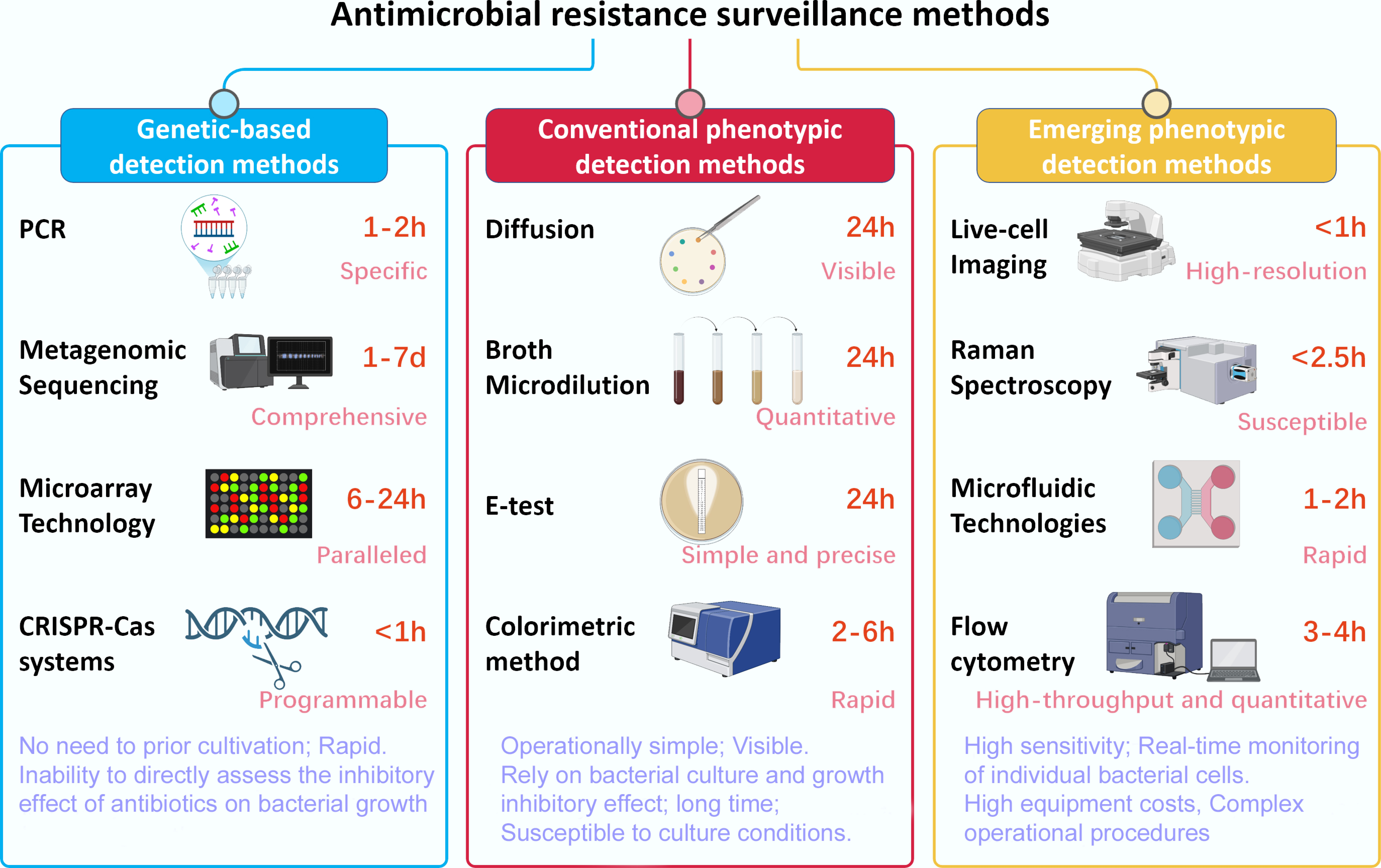
Figure 1.
The typical antimicrobial resistance surveillance methods include genetic-based, conventional phenotypic, and emerging phenotypic methods.
Table 1. Comparison of antimicrobial resistance surveillance methods
Technology Main principle Advantages and disadvantages Ref. PCR Detect antimicrobial resistance by amplifying resistance genes. Advantages: high sensitivity; detected rapidly (within 1–2 h)
Disadvantages: detecting the specific resistance genes; cannot assess antibiotic susceptibility[21,53−55] Metagenomic sequencing Analyze the total microbial DNA in a sample through high-throughput sequencing. Advantages: comprehensive without prior cultivation
Disadvantages: standardization challenges currently limited their clinical adoption[22,56,57] DNA microarray Immobilize specific probes on a chip and detect resistance genes in the sample through hybridization reactions. Advantages: high throughput (able to detect multiple resistance genes simultaneously); rapidness (suitable for clinical application)
Disadvantages: high cost; complex operation; and the risk of false positives (sample contamination)[40,58] CRISPR-Cas Utilize the RNA-guided nucleic acid recognition capability to detected ARGs with single-base resolution. Advantages: rapid (< 1 h), instrument-free detection suitable for point-of-care applications
Disadvantages: currently lack standardized protocols for widespread clinical implementation[42,43] Disk diffusion method The susceptibility is determined by the inhibition zone formed by the diffusion of antibiotics in the culture medium. Advantages: easy to perform; cost-effective; can test the susceptibility to multiple antibiotics simultaneously; suitable for routine clinical laboratories with limited resources
Disadvantages: partially automated; insufficient data support or poor detection performance for some bacteria; susceptible to environmental factors[23−26] Broth microdilution method The MIC is determined by the inhibition of bacterial growth at different concentrations of antibiotics. Advantages: accurately determine the MIC; has a high degree of standardization
Disadvantages: reagent requirements; high cost; complex operation; risk of false positives; risk of cross-contamination; the inability to distinguish between live and dead bacteria[37,38,45] E-test The MIC is determined by measuring the elliptical zone of bacterial growth inhibition created by a plastic strip containing a gradient concentration of antibiotics. Advantages: easy to operate; accurate in results; highly sensitive; suitable for a variety of bacteria, including slow-growing bacteria such as Helicobacter pylori
Disadvantages: not accurate enough for certain antibiotics (such as penicillin and ciprofloxacin); costly; has strict requirements for operating conditions[27−29] Colorimetric method Assess antibiotic susceptibility by detecting color changes caused by bacterial metabolism. Advantages: simple to use; low-cost; compatible with the standard microdilution method; does not require complex equipment
Disadvantages: low specificity; easily interfered with by other redox substances; long detection time (4–6 h)[46,47] Forward light scattering Detect changes in bacterial growth using laser scattering technology. Advantages: high throughput; rapid detection
Disadvantages: cannot distinguish between live and dead bacteria; high background noise[48,49] Real-time microscopy Conduct rapid detection using real-time microscopic imaging and fluorescence in situ hybridization (FISH) technology. Advantages: real-time monitoring (4–9 h); high degree of automation (the entire process from sample pretreatment to result analysis is automated)
Disadvantages: complex (requiring a high-precision microscope and an automated system); low throughput (only one sample can be processed at a time)[59−62] Live-cell imaging Monitor the growth and division of individual bacteria using microfluidic channels and microscopic imaging techniques. Advantages: rapid; high-throughput
Disadvantages: complex equipment and high cost equipment (high-resolution microscope and microfluidic system); high maintenance costs[33,62] Isothermal microcalorimetry Assess antibiotic susceptibility by detecting changes in heat produced by bacterial metabolism. Advantages: high sensitivity; free from matrix interference
Disadvantages: complex; data interpretation is relatively complicated[51,63] Mass-sensitive technology The technology based on microcantilevers assesses antibiotic susceptibility by detecting changes in bacterial mass. Advantages: high sensitivity (able to detect changes in individual bacteria); rapid (within 3–4 h)
Disadvantages: complex and high cost equipment (high-precision microcantilevers and vacuum systems); high maintenance costs[64,65] Electrical Assess antibiotic susceptibility by detecting changes in electrical properties such as current and voltage. Advantages: rapid (1–3 h); good portability
Disadvantages: low specificity; high background noise.[50,51] Motion tracking Assess antibiotic susceptibility by monitoring changes in bacterial movement using optical tracking technology. Advantages: rapid (within 30 min to 2 h); high sensitivity (detect changes at the single-cell level)
Disadvantages: complex and high cost equipment (high-precision microscopes and image processing algorithms); high maintenance costs[33,66] Raman spectroscopy Assess antibiotic susceptibility by detecting the Raman spectra of bacteria. Advantages: high sensitivity; rapid (within 30 min to 2.5 h)
Disadvantages: complex equipment; high background noise[30−32] Laser tweezers raman spectroscopy Capture individual bacteria with laser tweezers and perform Raman spectroscopy analysis. Advantages: single-cell analysis capability; background signal elimination
Disadvantages: complex equipment and operation[67,68] Fast Raman-assisted antibiotic susceptibility test Detect single bacterial metabolic activity in the presence of antibiotics, using Raman single-cell spectroscopy. Advantages: rapid (3 h) for urinary antibiotic susceptibility testing, enabling rapid clinical decision-making
Disadvantages: relies on sophisticated instrumentation and technical expertise[69,70] Fluorescence detection Detect the growth of bacteria by detecting metabolic activity. Advantages: high sensitivity (detect changes close to the single-cell level); rapid (within 3–4 h)
Disadvantages: high background noise; complex optical equipment and operation; poor performance on bacteria with low metabolic activity and the need for pure culture colonies[71−74] Microfluidic technologies Capture individual bacteria using microfluidic chips and conduct detection. Advantages: rapid (1–2 h); high throughput
Disadvantages: complex; strict limitations on sample volume[33,68] Flow cytometry Assess antibiotic susceptibility by detecting the scattered light and fluorescence of individual bacteria. Advantages: rapid detection; simultaneous analysis of multiple cell characteristics; high throughput; quantitative
Disadvantages: complex sample preparation; limited ability to analyze rare cells; high cost; complex equipment and operation[52,75] Genetic-based detection methods
-
PCR technology detects resistance by amplifying resistance genes, with high sensitivity, and the ability to complete detection within 1 to 2 h[21]. The key limitation of this method is the inability to directly assess the inhibitory effect of antibiotics on bacterial growth. Metagenomic sequencing analyzes the total microbial DNA in a sample through high-throughput sequencing, enabling comprehensive profiling of all genetic material (including bacteria, viruses, fungi, and resistance genes), without prior cultivation[22]. This culture-independent method is comprehensive for resistance surveillance but faces hurdles in clinical adoption due to bioinformatics complexity and longer turnaround times, compared to targeted PCR technology. Notably, human DNA is unavoidably present in metagenomic analyses of human microbiomes. Although standard protocols typically remove human DNA before public deposition, mitochondrial DNA (mtDNA) is often overlooked and persists in datasets. An exploratory analysis of 9,428 publicly available human metagenomes across 31 countries revealed that 1,817 samples (19.3%) contained human mtDNA, derived from stool, oral, and skin sources. The presence of human DNA can interfere with accurate quantitative microbiome profiling. More importantly, mtDNA constitutes personal genetic data, raising ethical and legal concerns. The WHO classifies mitochondrial genome data as part of the human genome, warranting similar safeguards. Thus, improving detection and removal of human-derived DNA is essential, and the balance between open data sharing and privacy protection must be carefully evaluated[39].
Compared to the above methods, microarray technology immobilizes nucleic acid probes on a chip to simultaneously detect multiple resistance genes via hybridization. This high-throughput approach is valuable for screening complex resistance profiles in a single assay, with turnaround times suitable for clinical use. However, challenges include high costs, technical complexity requiring skilled personnel, and potential false positives due to contamination or cross-hybridization[40]. On the other hand, emerging Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated proteins systems (Cas) utilize their RNA-guided nucleic acid recognition capability to detect ARGs with single-base resolution. Due to its ability to specifically target and cleave DNA sequences encoding ARGs, the CRISPR/Cas system has been developed into a novel gene-editing tool for the prevention and control of bacterial drug resistance. Compared to traditional nucleic acid detection technologies, the CRISPR-Cas system offers higher sensitivity and specificity, and its simplicity and portability facilitate on-site detection[41]. When integrated with isothermal amplification, these systems enable rapid detection (< 1 h)[42]. However, CRISPR/Cas9 faces challenges related to off-target effects, efficacy, and safety. These limitations call for effective methods to reduce off-target effects in targeted cells. Additionally, for broader acceptance, the system must address key areas such as cost and the development of standardized workflows that are validated across multiple centers during implementation[42,43].
Conventional phenotypic detection methods
-
Among the phenotypic drug susceptibility testing methods, conventional techniques usually rely on bacterial culture and the observation of growth-inhibitory effects. Although these methods are operationally simple and widely applicable in routine laboratory settings, they have a long detection time (4–24 h) and are susceptible to culture conditions. For example, the disk diffusion method relies on the diffusion of antibiotics in the culture medium to form an inhibition zone to determine bacterial susceptibility. Although cost-effective and capable of testing multiple antibiotics simultaneously[44], its accuracy can be compromised for certain fastidious or anaerobic bacteria (e.g., Bacteroides fragilis and Clostridium difficile) due to environmental interference[23−26]. The E-test integrates the advantages of both disk diffusion and dilution methods, using antibiotic gradient plastic strips to determine the MICs. Although it offers operational simplicity and relatively precise results, the downsides are its high cost (single-use strips) and stringent requirements for temperature, humidity, and a long incubation time[28,45]. Additionally, the colorimetric method assesses antibiotic susceptibility by detecting bacterial metabolic activity changes. Although simple and low-cost, this method has poor specificity, and is easily interfered with by the external environment[46,47].
Emerging phenotypic detection methods
-
The advantage of the emerging phenotypic detection methods lies in their high detection speed, making them suitable for clinical rapid testing needs. For instance, forward light scattering technology detects bacterial growth changes through laser scattering, reducing the detection time to within 3 h. However, if bacteria are not lysed, it is difficult to distinguish between live and dead bacteria, and the method is susceptible to background noise interference[48,49]. Real-time microscopy technology uses high-resolution microscopic imaging to analyze bacterial growth, offering a high degree of automation and the ability to complete detection within 4–9 h[40]. Besides, electrical detection technology assesses antibiotic susceptibility by detecting changes in current or resistance during bacterial growth, reducing the detection time to 1–3 h, but the equipment is relatively complex[50,51]. In addition, mass-sensitive detection technology uses microcantilevers to detect changes in bacterial mass, with high sensitivity that allows for the direct detection of individual cell changes[40]. On the other hand, motion tracking technology monitors changes in bacterial motility under the influence of antibiotics, with a short detection time of only 30 min to 2 h, but it requires a high-precision imaging system for data analysis[33].
Recently, emerging antibiotic susceptibility testing enables rapid bacterial response assessment without prolonged incubation, offering new possibilities for research and diagnostics. For instance, Raman spectroscopy assesses antibiotic susceptibility by analyzing molecular vibrations to detect antibiotic-induced biochemical changes in bacterial cells[30,32]. Capable of delivering results within 30 min to 2.5 h, Raman spectroscopy offers label-free, single-cell resolution analysis. However, it is susceptible to background interference and requires sophisticated spectral interpretation expertise[31]. Fluorescence detection technology assesses bacterial growth by detecting metabolic activity fluorescence signals, with some methods yielding results within 3 to 4 h, but its performance is limited for bacteria with low metabolic activity[40]. Notably, by integrating live-cell imaging technology, microfluidic systems, and microscopic imaging, this approach enables real-time monitoring of individual bacterial cells under antibiotic exposure[33]. This technology achieves remarkable detection speeds (≤ 1 h) while providing direct visualization of bacterial responses, and is regarded as an efficient means for clinical rapid testing.
Flow cytometry assesses antibiotic susceptibility by detecting the fluorescence characteristics of individual cells, with a fast detection speed and the advantage of high-throughput detection. This high-throughput method detects scattered light and fluorescence signals to evaluate bacterial viability and metabolic responses to antibiotics, offering advantages in speed and multiplexed testing capabilities[52]. Recent advancements have further enhanced its utility by incorporating nucleic acid probes that simultaneously detect resistance genes (e.g., mecA for MRSA, blaKPC for carbapenemases) alongside phenotypic responses. This dual detection approach bridges the gap between conventional growth-based antibiotic susceptibility testing and molecular diagnostics, enabling comprehensive resistance profiling within a single assay. However, the technique requires careful optimization of staining protocols and remains susceptible to background noise interference, which may affect signal interpretation. Despite these limitations, its ability to process thousands of cells simultaneously, while correlating genotypic and phenotypic resistance markers, makes it a powerful tool for both clinical microbiology and AMR surveillance. Despite significant advancements in detection speed, accuracy, and result visualization achieved by emerging technologies, their high equipment costs, complex operational procedures, and restricted scalability continue to limit widespread adoption in routine laboratory settings. Moving forward, it is essential to enhance the practicality and accessibility of these technologies through further innovation and cost-reduction efforts.
-
Breakpoints are predefined antibiotic concentration thresholds used to interpret MIC results[76]. Comparing the measured MIC to its breakpoint determines a bacterium’s susceptibility category for a given antibiotic. When the MIC value of a bacterium is less than or equal to the breakpoint, it is classified as a susceptible (S) strain; conversely, if the MIC value is higher than the breakpoint, it is classified as a resistant (R) strain[77]. Additionally, if the MIC value falls between the S and R breakpoints, the strain is classified as the Intermediate (I) category. These breakpoints were established by considering antibiotic pharmacokinetic (PK) and pharmacodynamic (PD) properties, clinical treatment success rates, and bacterial epidemiology[78], and were typically set and updated by professional bodies such as CLSI and EUCAST.
Multiple national organizations have established antimicrobial susceptibility breakpoints, including those in France, Sweden, Norway, the United Kingdom, the Netherlands, Germany, and the United States, each with distinct monitoring systems and priorities for clinical breakpoint (CBP) determination[79]. Currently, two international organizations serve as the primary authorities for breakpoint standardization: CLSI and EUCAST. These organizations continuously update breakpoint values and modify testing methods according to the evolving pathogens profiles and emerging clinical and laboratory data[20,80]. However, substantial variations exist in breakpoint settings, testing methodologies, and data interpretation across different standards, resulting in inconsistent antimicrobial susceptibility testing (AST) breakpoints among countries and regions. Such discrepancies not only hinder the comparability of global antimicrobial stewardship and resistance surveillance, but also create practical challenges for multinational research collaboration and clinical decision-making.
CLSI breakpoint standards
-
Established in 1968 as the National Committee for Clinical Laboratory Standards of America (and renamed in 2000), CLSI has become a globally recognized organization for providing standardized operating methods and judgment criteria for clinical laboratories[81]. Its AST standards have become one of the most important guidelines for clinical microbiology laboratories worldwide.
The committee of CLSI Antimicrobial Susceptibility Testing is composed of experts from various fields, including clinical microbiologists, infectious disease physicians, and pharmacists[81]. This committee is responsible for formulating, developing and validating methods, updating AST standards, and quality control to ensure consistency and reliability of testing results across laboratories[82]. The breakpoint setting process considers three critical parameters, including the wild-type cutoff (COWT)/epidemiological cutoff (ECOFF), pharmacodynamic cutoff (COPD), and CBP[83]. These parameters are determined by comprehensive analysis of global MIC data of clinical strains, the PK and PD characteristics of drugs, and by evaluating clinical research data to set breakpoint values for S, I, and R categories. CLSI places strong emphasis on integrating laboratory testing methods with clinical data, particularly through parallel comparisons of dilution and disk diffusion methods to refine the determination of intermediate values[83]. This standard was designed to ensure that breakpoints effectively guide the rational usage of antimicrobial agents in clinical practice. In addition, when the existing breakpoints no longer align with clinical outcomes or resistance patterns, the CLSI updates its standards to maintain accuracy in predicting antimicrobial efficacy, and support rational clinical decision-making[84].
EUCAST breakpoint standards
-
Established in 1997 by the European Society of Clinical Microbiology and Infectious Diseases, and the European Center for Disease Prevention and Control, EUCAST functions as the European regulatory authority for AST standardization, establishing evidence-based breakpoints for both existing and novel antimicrobial agents[79]. Its main responsibilities include setting and coordinating CBPs for antimicrobial agents, establishing ECOFF values, and developing and promoting standardized AST methods[85].
EUCAST employs rigorous scientific methods, and combines several critical parameters for breakpoint setting, including the ECOFF, PK/PD parameters, and CBP[80]. Firstly, EUCAST collected a large amount of MIC data from clinical strains, analyzed their distribution to determine the ECOFF, which distinguished wild-type strains from resistant strains. Secondly, it combined the PK/PD characteristics of the drugs to evaluate the effective concentration range of the drugs in the body. In addition, it assessed the clinical efficacy of different MIC values based on clinical research data, ensuring that the breakpoint setting reflects the actual needs of clinical treatment[86]. Finally, based on these data, EUCAST determined the breakpoint values for S, I, and R categories.
Discrepancies between CLSI and EUCAST in antimicrobial susceptibility standards
-
Despite the widespread adoption of CLSI and EUCAST standards, persistent differences between them pose significant challenges in practice. For instance, in international multicenter clinical trials, laboratories in different countries face data comparison difficulties when applying different breakpoint standards. Similarly, in global resistance monitoring programs, the absence of a unified standard hampers data integration and comprehensive analysis. For instance, there are differences in the breakpoint settings for ciprofloxacin between EUCAST and CLSI, leading to inconsistent resistance determination results for Escherichia coli under different standards[87−89]. Therefore, promoting cooperation between CLSI and EUCAST, and establishing a unified and authoritative MIC breakpoint system is crucial for improving the accuracy of global resistance monitoring, and the rationality of clinical antimicrobial use[36,83,89]. Specifically, the variation in MIC between CLSI and EUCAST, in terms of methods and evaluation guidelines, mainly manifests in the following aspects:
(1) Susceptibility classification. Before 2020, both CLSI and EUCAST used the 'Intermediate' (I) classification with consistent definitions. However, while CLSI retained the I category, EUCAST redefined it in 2020 as 'susceptible dose dependent', and introduced a new category, 'Area of Technical Uncertainty'[36]. This revision has led to certain strains being classified as I by CLSI while classified as R by EUCAST standards, resulting in higher resistance rates when applying EUCAST criteria[90]. For example, for a patient with a bloodstream infection caused by a Pseudomonas aeruginosa isolate exhibiting a ciprofloxacin MIC of 0.5 mg/L, CLSI classification as 'I' might support a high-dose treatment attempt, whereas the EUCAST 'R' designation would necessitate an immediate switch to an alternative antibiotic class. This discrepancy can directly increase the risk of initial treatment failure in the CLSI-guided approach or promote unnecessary broader-spectrum antibiotic use under EUCAST guidance, thereby increasing the risks of AMR.
(2) Discrepancies in numerical breakpoints. The differences in breakpoint values between organizations may lead to their varying susceptibility interpretations (Fig. 2). For instance, in determining the MIC breakpoint of anidulafungin against Candida albicans, CLSI defined the S breakpoint at ≤ 0.25 mg/L and the R breakpoint as ≥ 1 mg/L. In contrast, EUCAST set the S breakpoint at ≤ 0.03 mg/L, and the R breakpoint at > 0.03 mg/L[91]. This breakpoint discrepancy results in divergent clinical pathways for invasive Candida albicans infections: under CLSI guidelines, a patient would receive appropriate first-line anidulafungin therapy, whereas the same case under EUCAST standards would lead to unnecessary avoidance of anidulafungin and potentially less effective alternative antifungal regimens. A similar situation was observed with anidulafungin and micafungin against strains of Nakaseomyces glabrata (EUCAST S breakpoints: 0.06 and 0.03 for anidulafungin and micafungin, respectively; CLSI S breakpoints: 0.12 and 0.06 for anidulafungin and micafungin, respectively), Pichia kudriavzevii (EUCAST: 0.06 and no value; CLSI: 0.25 and 0.25), and Candida tropicalis (EUCAST: 0.06 and no value; CLSI: 0.25 and 0.24). These EUCAST criteria for susceptibility are consistently lower than the CLSI standards (Fig. 2), which can result in different interpretive outcomes for clinical practice.
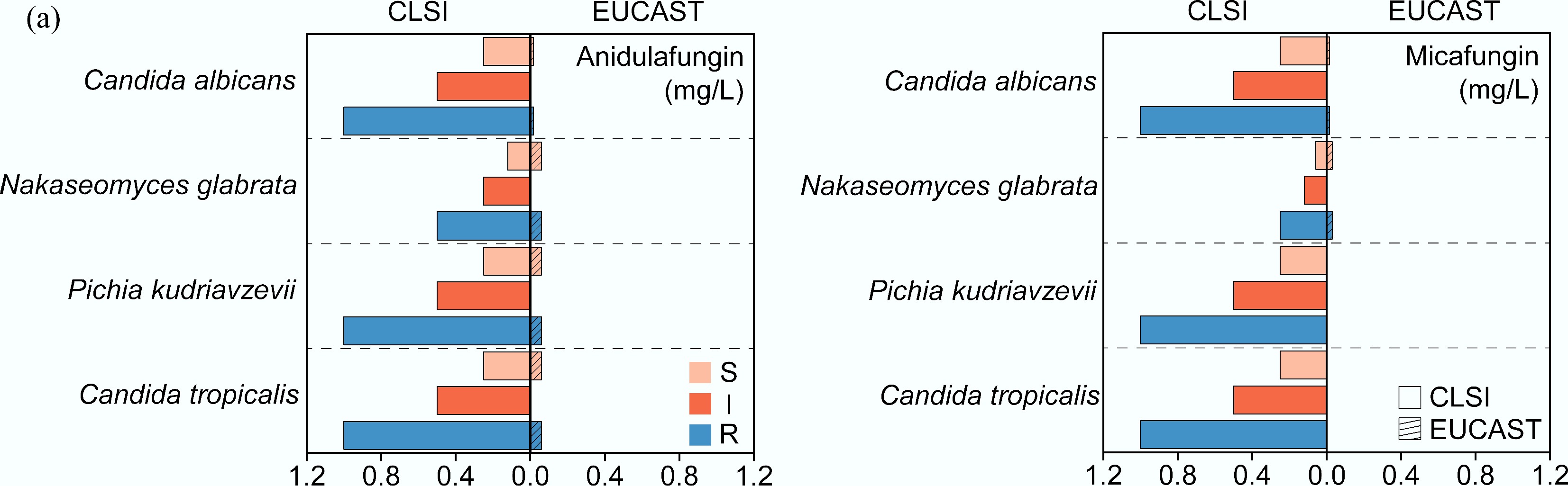
Figure 2.
Comparison of MIC breakpoints for (a) anidulafungin and (b) micafungin between CLSI and EUCAST. S: Susceptible, I: Intermediate, R: Resistant[91].
(3) Discrepant susceptibility classifications for identical bacterial strains. With changing antibiotic usage patterns and emerging resistance, both CLSI and EUCAST regularly update their breakpoints. The divergent breakpoint updates between the two institutes have led to the discrepant susceptibility classifications for identical bacterial strains. To bridge this gap, CLSI has updated several breakpoints, such as those for ciprofloxacin for Enterobacteriaceae and Pseudomonas aeruginosa, which brought them closer to those of EUCAST[89]. However, such updates may lead to the reinterpretation of historical data, and to a change in the resistance classification of certain strains. Consequently, when conducting MIC experiments or analyzing results, it is essential to ensure the use of the latest standards, and to clearly indicate the evaluation system, and its version used in the report.
(4) Multiple additional factors beyond breakpoint differences. For example, gram-positive and gram-negative bacteria exhibit significant differences in susceptibility and resistance to different classes of antibiotics. Alhumaid et al. found that gram-positive bacteria were resistant to ampicillin, cefoxitin, and doxycycline, while gram-negative bacteria were susceptible to these three antibiotics. Besides, gram-negative bacteria were resistant to tigecycline, meropenem, imipenem, and amikacin, while gram-positive bacteria were susceptible[92]. In addition, differences in bacterial strains, growth phases, physiological states, components of the culture medium, culture conditions, testing methods and instruments, as well as the physicochemical properties and action mechanisms of antibiotics, may all affect the results of susceptibility judgments[93]. These technical and biological factors collectively highlight the need for strict standardization of testing protocols and implementation of robust quality control measures across all AST programs to ensure result reliability and comparability.
(5) Impact on clinical decision-making. The inter-organizational breakpoint differences created substantial challenges in MIC data comparability across laboratories (Table 2). For instance, when classifying 428 Escherichia coli strains for amoxicillin-clavulanate susceptibility, CLSI criteria categorized 55.6% as S category, 24.5% as I category, and 19.9% as R category. In contrast, EUCAST standards resulted in a lower percentage of strains classified as S (47.7%) category and a higher proportion as R (52.3%) category. Such discrepancies increased the risk of antibiotic misuse in clinical practice: (1) a false S classification would lead to the use of ineffective drugs and subsequent treatment failure; and (2) a false R classification might result in the unnecessary avoidance of narrower-spectrum agents and increased use of broader-spectrum alternatives, thereby accelerating AMR. Furthermore, for strains with I classification or borderline MICs, CLSI would recommend increasing the drug dose to achieve therapeutic efficacy, while EUCAST usually suggests alternative antibiotic therapy[90]. These discrepancies not only affect the treatment of an individual patient but also impact a hospital's antibiotic management strategies, including antibiotic usage policies and resistance alerting[89].
Table 2. Susceptibility analysis of different antibiotics to bacteria based on CLSI and EUCAST
Antibiotic Strain and the test number CLSI (%) EUCAST (%) S I R S I R Amoxicillin-clavulanate Escherichia coli (428) 55.6 24.5 19.9 47.7 − 52.3 Klebsiella pneumoniae (208) 67.3 10.6 22.1 64.4 − 35.6 Ciprofloxacin Escherichia coli (428) 50.5 3 46.5 31.3 9.6 59.1 Klebsiella pneumoniae (208) 72.6 13.5 13.9 47.6 14.9 37.5 Pseudomonas aeruginosa (78) 85.9 3.8 10.3 71.8 0 28.2 Gentamicin Escherichia coli (428) 58.4 0 41.6 55.1 2.8 42.1 Klebsiella pneumoniae (208) 76.4 0 23.6 73.6 2.9 23.6 Pseudomonas aeruginosa (78) 76.9 0 23.1 75.6 0 24.4 Ceftriaxone Escherichia coli (428) 41.8 0.2 57.9 40.4 1.4 58.2 Klebsiella pneumoniae (208) 68.8 0 31.3 64.9 3.8 31.3 The number of strains refers to the total count of bacterial isolates tested for their susceptibility to antibiotics[87−89]. A higher percentage in the S category indicates a greater number of susceptible isolates among the tested strains, while a higher percentage in the R category indicates a greater number of resistant isolates. S: Susceptible, I: Intermediate, R: Resistant. Therefore, achieving global harmonization of CLSI and EUCAST breakpoint standards represents an urgent priority for advancing AMR management. Future collaboration should focus on four critical dimensions: first, aligning the interpretive criteria for S, I, and R categories to support clinical decision-making; second, unifying drug-strain-specific susceptibility categories and epidemiological cut-off values; third, standardizing technical methodologies covering strain selection, culture protocols, and MIC detection systems; fourth, establishing a unified evidence-based framework for breakpoint derivation that integrates PK/PD data with clinical outcomes. Such alignment will transform AST into a universally applicable tool that enables seamless cross-regional data comparison, strengthens global antibiotic stewardship programs, and establishes a solid foundation for detecting emerging resistance patterns in both clinical and environmental settings. Moreover, this convergence will enhance public health capacity to combat AMR through more precise therapeutic decisions and reliable global surveillance.
-
As antibiotic resistance continues to increase, rapid, high-throughput, and automated surveillance technologies are becoming essential for future development. Although traditional surveillance methods still hold significant value in resource-limited settings, their limitations are becoming more apparent. The emerging technologies (such as optical, electrical, and molecular biological detection technologies) provide new insights for solving this problem. However, the widespread application of these technologies still faces challenges, such as high equipment costs, complex operations, and strict requirements for environmental and sample conditions. Therefore, it is necessary to select surveillance methods reasonably based on factors such as the purpose of detection, laboratory conditions, and sample types to ensure the accuracy and reliability of the results in practical applications. For example, in clinical rapid diagnosis, fluorescence detection and electrical detection technologies may have more advantages, while traditional disk diffusion and dilution methods were more suitable in resource-limited environments.
As the most authoritative organizations for setting AST standards globally, the differences in breakpoint systems between CLSI and EUCAST had a profound impact on resistance monitoring and clinical antibiotic use. CLSI placed more emphasis on the combination of laboratory testing methods and clinical data, highlighting the setting of intermediate values to provide more treatment flexibility. In contrast, EUCAST mainly set breakpoints based on PK/PD data, usually without intermediate values, and emphasized the efficacy of drugs at the highest recommended doses. This difference lead to significant discrepancies in the susceptibility classification of the same bacterial strain under different standards, thereby affecting clinical decision-making and antibiotic usage strategies. Moreover, differences between CLSI and EUCAST in operational procedures such as medium selection, inoculum preparation, quality control strain selection, and incubation time further exacerbated the inconsistency of results. Therefore, there is an urgent global need to establish a unified and authoritative breakpoint system to enhance the comparability of resistance monitoring and the rationality of clinical antibiotic use.
Future research should focus on addressing the following issues. First, on the development of more efficient, low-cost, and user-friendly antimicrobial surveillance technologies tailored to diverse application scenarios. Key directions include high-throughput automated systems for rapid susceptibility profiling in clinical samples, as well as highly sensitive biosensors and field-deployable assay technologies, capable of sensitive detection of ARB and ARGs in environmental matrices, such as water and soil systems. Second, on strengthening cooperation and coordination between CLSI and EUCAST to narrow the differences in breakpoint systems and to promote the standardization of global resistance monitoring and antimicrobial management. By integrating multidisciplinary research findings (such as genomics, metabolomics, and pharmacokinetics), in-depth studies on the formation mechanisms and transmission pathways of antibiotic resistance should be conducted to provide a scientific basis for the development of new antimicrobial agents and the formulation of resistance control strategies. In addition, with the continuous development of artificial intelligence (AI) technologies, their applications in resistance monitoring and surveillance technologies are also worth further exploration. For example, machine learning can be utilized to analyze complex datasets (such as genomic sequences) to achieve rapid and high-accuracy pathogen identification, and resistance profiling. Moreover, AI-driven models can integrate large-scale clinical and environmental datasets to predict the emergence and dissemination of resistant strains across different ecological settings. Finally, significant gains can be made through workflow optimization, where implementing AI algorithms can streamline the entire surveillance process, from automated data entry and quality control, to the interpretation of results, thereby significantly reducing time and resource costs.
In summary, the surveillance technologies for ARB and the MIC breakpoint systems are key elements in current research and clinical practice. Although existing technologies have made significant progress in detection speed, sensitivity, and specificity, their widespread application still faces many challenges. Meanwhile, the impact of differences in CLSI and EUCAST breakpoint systems on resistance monitoring and clinical antibiotic use cannot be ignored. In the future, continuous efforts are needed in technology optimization, standard unification, and multidisciplinary cooperation to address the increasingly serious issue of AMR, and provide reliable data support for control measures and policy decisions.
-
The authors confirm their contributions to the paper as follows: Moran Tang: conceptualization, methodology, software, writing – review & editing. Zehui Wang: conceptualization, methodology, software, writing – original draft. Hong Zhu: methodology, software. Nyuk Ling Ma: software, writing – review & editing. Zhenzhen Yang: methodology, software. Yunlong Tian: methodology, software. Hongna Li: conceptualization, methodology, writing – review & editing, supervision, funding acquisition. All authors reviewed the results and approved the final version of the manuscript.
-
The datasets generated during and/or analyzed in the current study are available from the corresponding author upon reasonable request.
-
This research was funded by the National Key Research and Development Program (Grant No. 2024YFD1700602), Ningxia Key Research and Development Program (Grant No. 2023BCF01049), the Top-Notch Young Talents Program of China, the Agricultural Science and Technology Innovation Program of China (Grant No. CAAS-CFSGLCA-IEDA-202302), and the Central Public-interest Scientific Institution Basal Research Fund (Grant No. Y2025C30). The authors declare no competing financial interest.
-
The authors declare that they have no conflict of interest.
-
Minimum inhibitory concentration (MIC) remains the gold standard for antibiotic resistance assessment.
Current antimicrobial resistance surveillance relies on genetic-based and phenotypic methods.
Breakpoint discrepancies between authoritative bodies cause inconsistent susceptibility classification.
A globally unified MIC breakpoint system is urgently needed.
-
# Authors contributed equally: Moran Tang, Zehui Wang
Full list of author information is available at the end of the article. - Copyright: © 2026 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Tang M, Wang Z, Zhu H, Ma NL, Yang Z, et al. 2026. Antimicrobial resistance surveillance in the natural environment: standardization of minimum inhibitory concentration breakpoint. New Contaminants 2: e003 doi: 10.48130/newcontam-0025-0023
Antimicrobial resistance surveillance in the natural environment: standardization of minimum inhibitory concentration breakpoint
- Received: 22 October 2025
- Revised: 17 December 2025
- Accepted: 26 December 2025
- Published online: 23 January 2026
Abstract: The widespread dissemination of antimicrobial resistance (AMR) in the natural environment poses a severe threat to global public health, necessitating reliable surveillance methods, and robust monitoring systems to combat it. This work comprehensively reviews current AMR surveillance methods, including genetic-based and phenotypic methods. Genetic-based methods (e.g., polymerase chain reaction, CRISPR-Cas) offer rapid, highly specific identification of resistance genes but cannot identify novel mechanisms or recognize phenotypic resistance profiles. Conventional phenotypic methods (e.g., disk diffusion, broth dilution), although widely used due to their relatively simple-operation and cost-effectiveness, are time-consuming and susceptible to environmental factors. Emerging phenotypic methods (e.g., Raman spectroscopy, flow cytometry) offer fast speed, high sensitivity, and high throughput for AMR surveillance. However, they require sophisticated and costly instrumentation and technical expertise. Among these methods, phenotypic techniques are employed to determine the minimum inhibitory concentration (MIC) of antibiotics, which serves as the gold standard for assessing antibiotic resistance. The interpretation of MIC values is guided by breakpoint systems established by authoritative organizations such as the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). However, differences in breakpoint thresholds, testing methodologies, and data interpretation between these systems lead to substantial discrepancies in classifying the susceptibility of the same bacterial strain. These discrepancies could cause inconsistent treatment recommendations regarding dose escalation or alternative antibiotic choices, ultimately affecting both antimicrobial therapy and stewardship policies. This article calls for a unified MIC breakpoint system to standardize global resistance data, and guide antibiotic use. The development of cost-effective, user-friendly surveillance technologies for AMR needs to be developed via multidisciplinary collaboration.