-
Cassava (Manihot esculenta Crantz), belongs to the family Euphorbiaceae, and is an important staple crop for about 800 million people worldwide[1,2]. It is a key commodity for food processing industries[3]. It is mostly grown by smallholder farmers in more than 100 tropical and subtropical countries. Cassava contains a high proportion of starch (70%−85% of the root dry matter content)[1] and, it is the second most traded source of starch after maize[2,4]. Cassava originates from the southern Amazon region[5] and was introduced to Africa by the Portuguese in the 16th century. Its cultivation was expanded in the 20th century when it emerged as an important food crop in India, Indonesia, the Philippines, and sub-Saharan Africa (SSA)[2,6−8]. Currently, it is grown almost exclusively in tropical and sub-tropical regions with various names. For example, it is called mandioca in Brazil, ketela pohon in Indonesia, yuca in many Spanish-speaking countries, akpu in Nigeria, sắn in Viet Nam, mihogo in Kenya[2] and imyumbati in Rwanda.
In Africa, cassava is a food security crop due to its resilience to grow under harsh conditions including poor soils, drought, and the ease with which it can be propagated vegetatively[9]. In East Africa, cassava storage roots can be prepared in various ways. Sweet varieties are eaten raw, boiled or processed into flour. Hence, various recipes such as ugali, porridge, alcohol beverages and bread are prepared from cassava[10]. Cassava leaves are consumed as an important vegetable in some countries[11] including Rwanda and the Democratic Republic of Congo[12,13]. The higher micronutrient and protein contents of cassava leaves can address nutritional deficiencies associated with the consumption of cassava storage roots only[14−16].
The worldwide cassava production was 302.6 million tons on 28.2 million hectares in 2020. More than 50% of this production (193.6 million tons) occurs in Africa. Asia and Latin America produce 27% (81.8 million tons) and 8.2% (25 million tons) respectively[17]. Under optimal conditions, the potential yield for cassava can reach 90 tons of fresh roots per hectare[18]. In east Africa, cassava productivity under optimal conditions varies between 50−60 tons per hectare[19,20]. The realized average yield in Africa is 8.6 tons per hectare under small-scale farmers. The use of improved varieties and good agriculture practices boosts yield reaching up to 20.8 tons per hectare[20] or to 24 tons per hectare[21].
The average cassava yields in Africa remain relatively low compared to those of the Asian continent, where the average yield is about 21.8 tons per hectare[17]. This yield difference is due to various abiotic (inadequate rainfall, low input use, low soil fertility) and biotic (weeds, pests and diseases) factors and, this is exacerbated by sub-optimal management practices[20,21]. Another reason for the low productivity of cassava in Africa is that it is often grown in intercropping systems with two or more crops[22]. For instance, cassava yield can be reduced by nearly 60% under drought conditions. In fact, water stress during the first six months after planting (MAP) or during the entire cropping season is disastrous because it decreases cassava storage root initiation and hence reduces root yield[20]. Several parameters, including leaf longevity and stomatal conductance, can explain the contrasting performance of drought-susceptible and drought-resistant genotypes[23]. A strong positive correlation between leaf retention and yield as well as a positive correlation between leaf longevity and yield were reported[23,24]. An increase in leaf retention improves cassava total biomass production and accumulation into the roots, which in turn, results in a higher root yield[24]. In addition, higher stomatal conductance and delayed stomatal closure can result in high yields in drought-tolerant genotypes, while the opposite can lead to low yields in drought-susceptible genotypes[23]. Another notable example is that poor weed management during three MAP was reported to cause cassava yield reductions of 50%−65%[20].
Viral diseases are the leading causes of this low productivity[25]. Two viral diseases namely cassava mosaic disease (CMD) caused by Geminiviruses and cassava brown streak disease caused by Ipomoviruses, are considered the main biotic constraints to cassava production in Africa[26,27]. The annual cassava monetary losses caused by both CMD and CBSD have been estimated to be over USD
${\$} $ ${\$} $ Importantly, leaf and root symptoms of CBSD are not necessarily correlated[32]. For the farmers, root symptoms are only visible at harvest when damage has already occurred. These 'silent' symptoms of CBSD also complicate breeding efforts and the ability to produce disease-free planting material. CBSD was first reported in the 1930s from north-eastern Tanzania[35]. Subsequent surveys indicated that the disease was widespread in coastal East Africa[36]. Based on the field observations, it was reported that the virus causing the disease could not spread and establish at altitudes above 1,000 m above sea level. The first reports of CBSD spread into higher altitude zones of East Africa were made in 2004[29] as new occurrences of CBSD were observed in southern Uganda. CBSD was subsequently reported in several countries from the Great Lakes region of East/Central Africa[31]. CBSD is caused by two distinct virus species namely Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV). These are referred to, collectively, as cassava brown streak ipomoviruses (CBSIs)[37−39]. Both viruses are positive sense single-stranded RNA (ssRNA) belonging to the genus Ipomovirus, family Potyviridae[40,41]. CBSIs are transmitted by a whitefly vector − Bemisia tabaci (Genn) but their dissemination also occurs through infected cuttings[42,43]. The typical symptoms of CBSD (Fig. 1a−d) are interveinal leaf chlorosis without distortion of the lamina, brown lesions on green stems, necrosis of leaf scars and terminal die-back, root necrosis and stunting of plants especially in susceptible varieties[44]. The disease has only been reported in African countries (Table 1). Concerted efforts are needed to prevent its spread, which will likely continue to all main cassava-growing areas of Africa and probably to Southeast Asia[3,31].

Figure 1.
Symptoms of cassava brown streak disease. (a) Leaf chlorosis without distortion of the lamina and brown lesions on a green stem. (b) Storage root constrictions. (c) Mild necrosis of cassava storage roots. (d) Severe necrosis of cassava storage roots.
Table 1. List of countries where cassava brown streak disease is reported.
The use of resistant varieties and the distribution of disease-free planting materials are sustainable strategies to control CBSD[2,53]. Although recently identified sources of CBSD resistance[54] might offer new opportunities to mitigate CBSD in the field, the sources of CBSD resistance or tolerance used so far in breeding programs have not been satisfactory. Because CBSD-tolerant varieties can support relatively important titers of CBSIs, CBSD resistance is usually preferred in breeding programs as it reduces the presence of virus inoculum in the fields and limits the probabilities of virus evolution.
Cassava brown streak disease breeding started at Amani Research Station (ARS), Tanzania in the 1930s[55,56]. Early conventional breeding studies showed that generating cassava with high levels of resistance against CBSD was possible. CBSD resistance was introgressed into cultivated cassava through crosses with Manihot glaziovii Müll. Arg. followed by repeated backcrossing[56]. This allowed the development and deployment of CBSD-resistant clones in East African countries affected by CBSD since 1956[44]. The extensive use of the hybrid 46106/27 (a back-cross three generation [BC3] between Manihot glaziovii Müll. Arg. and Manihot esculenta Crantz) in breeding programs illustrates the importance of cross breeding with Manihot glaziovii Müll. Arg. as a source of CBSD resistance. The hybrid 46106/27 is also named Kaleso in Kenya and Namikonga in Tanzania and it remains an important parental line in several conventional breeding programs for virus resistance[57,58]. Notably, CMVs and CBSIs can be detected in Manihot glaziovii Müll. Arg. and other wild plant species[59,60], implying that Manihot glaziovii is no longer the only Manihot host for CBSIs in Africa. All Manihot species originate from the Americas, and it can therefore be postulated that they have never been challenged by CMD or CBSD in their evolutionary history. It also indicates that in Africa, CBSIs could have evolved over time to infect Manihot glaziovii Müll. Arg. that has been reported as a source of resistance to those viruses.
Early breeding efforts were successful in achieving resistance to CBSD. However, the resistance was not durable and overcome by the CBSIs. Genotypes known to possess durable resistance have become relatively susceptible to these viruses, even though virus multiplication and symptom expression are limited compared to the susceptible genotypes. The most illustrative example is the breakdown of CBSD resistance in the hybrid Kaleso/Namikonga[58]. CBSD resistance breakdown was also reported in NASE 14, a CBSD-tolerant variety widely grown in East African countries since its release in 2011. This genotype was recently reported to have high and severe leaf and root incidences of CBSIs in Uganda[61]. Recent research also suggests that deployed cassava varieties could exert selection on CBSIs as widely distributed cassava varieties are potentially associated with specific UCBSV haplotypes[62]. The molecular mechanisms for the resistance breakdown in previously resistant / tolerant varieties remain to be elucidated[63]. Notably, CBSIs have also been detected in M. glaziovii Müll. Arg. which previously served as a putative source of CBSD resistance[59,60]. Conventional breeding should be complemented with new plant breeding technologies (NPBTs) in speeding and scaling up cassava breeding efforts towards CBSD resistance. In this context, the search for other sources of resistance is vital[53,54,64,65]. Given the endemic presence of CBSIs in eastern Africa, it is also key to support cassava breeding initiatives in eastern and southern Africa[66]. This review provides an update on employed breeding strategies to fight CBSD and discusses the potential of NPBTs as an additional key tool for effective CBSD management in Africa.
-
Cassava is a highly heterozygous and commonly vegetative reproducing crop using stem cuttings or tissue culture systems[67]. Its vegetative propagation promotes the development and selection of homogenous clones and enhances trait heritability and selection response. However, cassava has a monoecious flowering system and produces true seeds through self- or cross-fertilization. Typically, cassava is a cross-fertilizer due to protogyny, that is, the stigma is receptive about two weeks before the pollen grain is released from the same flower[68]. Protogyny enhances natural out-crossing or poly crosses and the development of half-sib progenies, while controlled crosses involving selected parents allow the development of full-sib progenies[3,69]. During controlled pollinations, the mature pollen is collected and stored for a few hours while waiting for the opening of female flowers which usually happens later during the day. It is also practical to cover female flowers on the day of anthesis to avoid any contamination with unwanted pollen[70]. Cross-fertilization induces genetic diversity and allows the selection of new recombinants[71−73]. Self-fertilized seeds are produced due to the synchronization of stigma receptivity and pollen release from flowers of the same plant or from flowers of different plants of the same genotype[57]. Continuous and controlled selfing enables the development of homozygous and homogenous individuals for future cross-breeding or genetic analysis. However, cassava flowering is under the influence of genotype × environment interaction and, therefore, it is sometimes challenging to cross certain genotypes[74].
-
Although the initial hybridization work implemented at ARS, Tanzania in the 40's and 50's has provided valuable CBSD resistant breeding material, the breakdown of the CBSD resistance in the last decades calls for immediate actions to mitigate CBSD in Africa, from the identification and exploitation of new sources of resistance to the implementation of phytosanitation (Fig 2).
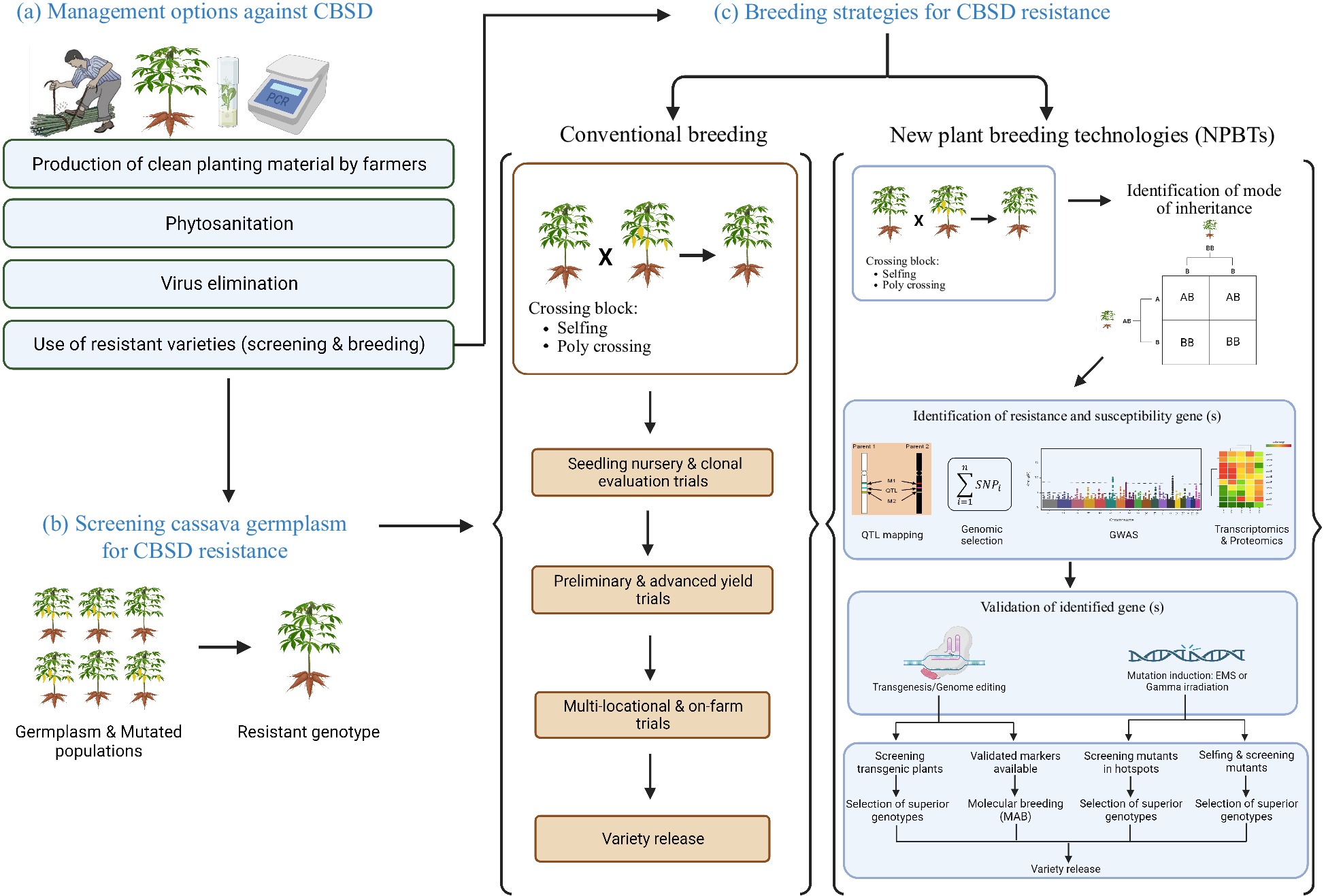
Figure 2.
Illustration of management and breeding strategies used to mitigate CBSD in Africa. (a) Various CBSD management options were used. Those are the production of clean planting material by farmers, phytosanitation, virus elimination using tissue culture, thermotherapy and chemotherapy and the use of resistant varieties. (b) Screening cassava germplasm and mutated population for CBSD resistance can be carried out either in the fields (CBSD hotspots) or in controlled environments where grafting method or infectious clones can be used. (c) In conventional breeding, a resistant genotype is crossed with a susceptible one and GCA and SCA for CBSD resistance are evaluated in progenies. Various genomic tools and NPBTs for CBSD resistance were used. Crossing of an identified resistant parent with susceptible farmer-preferred genotype is followed by a determination of the mode of inheritance, the candidate genes, and then validation of candidate genes. Validated markers are used as inputs in molecular breeding. After gene validation, field experiments are conducted to screen and select adapted superior genotypes for release. Conventional breeding is lengthy because it can take 8−10 years. Using NPBTs can help to shorten this cycle to few years: starting with mapping population development for marker discovery (2 years), marker development and validation (1 year) and field trials (2 years), the breeding cycle can take about 5 years. Once the markers are available, this cycle can be even shorter i.e., 3−4 years. This figure was created with BioRender.com.
Some local tolerant varieties have been identified and used as an option to manage CBSD in Tanzania and Mozambique (Fig. 2a). For instance, a local genotype 'Nanchinyaya' that showed CBSD tolerance was successfully used in southern Tanzania[75].
Phytosanitation, which involves using healthy planting materials and practicing roguing[76] represents a valuable approach for CBSD management (Fig. 2a). It encompasses: 1) The use of clean planting materials obtained after tissue culture, regular virus indexing for pre-basic generation seed supplemented by a consistent roguing of diseased plants in the field; 2) Collective interventions aimed at urging farmers living in the same locality to collaborate in implementing phytosanitary actions[77]. The effectiveness of community phytosanitation for managing CBSD was assessed in two communities of Tanzania (Mkuranga and Chato districts) from 2013 to 2016. In both target communities, large reductions in CBSD incidence as well as cassava yield increase were recorded[37]. The success of this approach calls for the rapid implementation of phytosanitation measures in integrated cassava virus management programs in places where CBSD remains a major constraint to cassava production.
Region-wide, large scale measures have also been implemented to restrict the spread of CBSD across the African continent. Movement of planting materials across the regions can be controlled through strict quarantine measures in order to make sure that the disease is not brought beyond its confined area[78]. Therefore only tissue cultured and virus tested materials are recommended while exchanging germplasm between countries[79]. This measure will continue to be essential in order to prevent the spread of CBSD in West African countries where no CBSD occurrence has been reported so far.
Given the costs associated with the distribution of disease-free planting material, the sustainable and cost-effective option for managing CBSD in the African context remains the use of resistant varieties by farmers[53]. In addition, farmers should use stems and cuttings from plants whose roots did not show symptoms to prevent disease dissemination and resistance breakdown. Therefore, there is an urgent need to develop CBSD-resistant varieties as a collaborative regional effort and to promote good practices for planting material. Breeding programs should not only target CBSD resistance, but also include on additional farmer- and consumer-preferred traits in order to ease the adoption of new varieties.
-
Screening for natural resistance in the cassava germplasm represents another important approach to provide the breeder's community with new sources of CBSD resistance. Usually, CBSD screening experiments are conducted in the field (virus hotspots) or in controlled conditions (screenhouses) (Fig. 2b). Genotypes are then evaluated for leaf and root necrosis symptoms (incidence and severity) as well as for the viral load via reverse transcription quantitative real time polymerase chain reaction (RT-qPCR).
Available protocols for CBSD resistance screening
-
Methods and techniques for studying viral diseases are essential in order to investigate plant host resistance[80,81]. Various protocols have been developed in order to screen cassava germplasm for CBSD resistance. Some protocols were applied in the fields, while others were used in the greenhouse and complemented by laboratory analysis for virus detection and quantification.
CBSD resistance screening under field conditions
-
Field screening activities for CBSD resistance are generally conducted in disease hotspot areas. Field experiments are typically established in a randomized complete block design (RCBD) with replications for at least two seasons. A susceptible genotype is usually included in the experiment and disease spreader rows are established inside and around the experiment to increase the inoculum pressure and ensure that all test genotypes are equally exposed to CBSD inoculum[58,61,82−84]. Cassava screening for resistance against CBSIs remains complicated and time consuming, not only because of random and slow virus infections, but also the reliance on root necrosis evaluation as an ultimate evidence of cassava plant resistance[64]. Therefore, it is recommended that root assessment takes place at 12 MAP because root necrosis symptoms become more visible and severe as cassava plants mature[36,44,85].
Disease assessment is carried out by evaluating CBSD incidence and severity on both leaves and roots. Incidence and severity are repeatedly assessed at 2−3 month intervals[58,84]. For CBSD severity leaf assessment, a scale of 1−5 is used where 1 corresponds to no apparent symptoms and 5 corresponds to defoliation with stem lesions and pronounced dieback. Root severity assessment is accomplished at harvest (12MAP). The root severity scale of 1 to 5 is used (1) no apparent symptoms, (2) less than 5% of root necrosis, (3) about 5%−10% of root necrosis, (4) about 10%−25% of root necrosis and mild root constriction, (5) greater or equal to 25% of root necrosis and severe root constriction[86]. Because cassava genotypes that are free of CBSD leaf symptoms do not necessarily have roots free of CBSD necrotic symptoms, a complete evaluation of CBSD resistance usually requires the inspection of the storage roots at the end of the growing cycle.
CBSD resistance screening under greenhouse conditions
-
A number of protocols to screen cassava germplasm for CBSD resistance were implemented in controlled conditions (greenhouses and laboratories). Chip bud-grafting method was developed for CBSIs inoculation in the greenhouse. Axillary buds from six to eight week old CBSV-infected rootstock plants carrying single or mixed CBSV and UCBSV infections are grafted into test plants[80]. The use of the most virulent isolate CBSV-Mo83 (DSMZ PV-0949, GenBank accession FN434436) with the bud grafting method is important because it allows significant shortening of the screening duration as CBSD symptoms appear at one month after grafting[64].
Top cleft-grafting and side grafting methods can also be used to screen cassava genotypes for CBSD resistance under greenhouse conditions (27 °C, 16 h of light, 60% humidity)[81]. The top cleft-grafting method whereby a scion is inserted into infected rootstocks remains very effective as it maximizes the infection rates and avoids escapes from infection. For instance, using this method, breeding lines KBH 2006/18 and KBH 2006/26 remained asymptomatic for 16 weeks after being inoculated with U/CBSV while susceptible genotypes displayed symptoms at 4 weeks after inoculation[87]. The top-grafting method has also been used for screening Ghanaian cultivars to assess the susceptibility of cassava germplasm used in Western Africa and not yet exposed to CBSD pandemics in the field[88].
Although the grafting methods have enabled screening of CBSD resistance, they remain time-consuming and sometimes display suboptimal infection rates. Infectious clones have been used as an important tool for virus-resistance screening studies[89,90]. Viral infectious clones contain genome copies of viral pathogens in bacterial plasmids that provide a large inoculum to initiate plant infection. They are delivered to plants by physical introduction with the help of abrasives or biolistic devices using viral RNA transcripts or plasmid DNA, or by using Agrobacterium-mediated plant inoculation. U/CBSV infectious clones have been successfully used to inoculate Nicotiana benthamiana species using Agrobacterium-mediated plant inoculation[91−93]. However, there has been limited success in infecting cassava plants by agroinoculation with the CBSV infectious clones and grafting has so far remained the method of choice for CBSD resistance screening experiments[92]. Therefore, there is a need to develop infectious clones of CBSIs effective at establishing CBSD in cassava as well as to investigate the biological factors preventing effective inoculation with clones of CBSIs in cassava. Cloning strategies and inoculation methods might require further optimization in order to achieve good infectivity in cassava[90].
Screening efforts to find CBSD resistance in the cassava germplasm
-
Since the re-emergence of CBSD, efforts have been launched to screen the cassava germplasm for natural resistance/tolerance to CBSD. The screening activities have been initially performed on cassava genotypes used by farmers or in breeding programs in Africa. In recent years, the screening activities have been expanded to the whole cassava germplasm, including genotypes from Southern America[54].
In Africa, several resistant and tolerant genotypes were identified amongst evaluated genotypes[29,82,84,88,94,95]. These resistant / tolerant genotypes could be used by farmers to reduce CBSD losses or by breeding programs as good sources of resistance against CBSD. Surprisingly, though CBSD is not yet reported in West African countries, the fact that nearly all Nigerian genotypes have shown susceptibility to CBSD[82,88] is alarming and, it informs West African cassava breeders that their cultivars lack resistance to this disease and therefore re-emphasizes that they should avoid importing cassava germplasm from East African countries. It should be remembered that the movement of cassava planting materials from West Africa to East Africa is officially allowed but the opposite is forbidden to avoid any introduction of CBSD in West Africa[77].
Although various screening efforts are ongoing to find CBSD resistance in African germplasm, it was realized that resistance present in Africa is not sufficient to cope with the disease, therefore additional sources of virus resistance were searched in cassava collections and cassava germplasm prevalent outside Africa. In this regard, 238 cassava lines from America (CIAT) were screened for CBSD resistance under greenhouse conditions using the CBSV-Mo83 isolate with the bud-grafting method. Most of the accessions were susceptible, but, interestingly, seven genotypes remained healthy with no symptoms and no virus detection in both leaves and roots[54]. This resistance found in American genotypes is justified by the fact that viruses (U/CBSVs) could not replicate and are only restricted to the phloem companion cells[65]. After greenhouse experiments, extensive field trials were conducted in Africa, and promising results were obtained. In fact, three genotypes (COL 40, PER 556 and COL 2182) exhibited high levels of resistance to U/CBSV and they were considered as immune[65]. These genotypes will serve as another important source of resistance to CBSD that breeders in NARS (National Agricultural Research System) can use in their future breeding programs. Once crossed with highly tolerant genotypes (e.g: KBH 2016B/504), these genotypes can provide significant resistance to U/CBSV and hence, contribute to the efforts towards mitigating CBSD impact in Africa. Nevertheless, it was observed that several CBSD-resistant or immune genotypes are susceptible to CMD. Therefore, additional breeding efforts will be required to combine resistances to both viral diseases.
-
Securing resistance against CBSD, is a prerequisite for sustainable cassava production in Africa. Thus, developing CBSD resistant varieties will help to grow a healthy and productive crop while preventing the spread of the virus in regions that are not yet affected on the continent (West Africa)[65].
Conventional breeding strategies for controlling cassava brown streak disease
-
Conventional breeding is the most widely used approach for developing new cassava varieties[67]. In this process, genetic variability is generated by crossing two flowering plants and, large numbers of plants are advanced until desired genotypes are identified and selected. Then, selected genotypes undergo performance evaluation before the official release to farmers (Fig. 2c)[96].
In a study to evaluate the importance of general combining ability (GCA) and specific combining ability (SCA) and inheritance of relevant traits, Zacharias and Labuschagne crossed five parental genotypes using a full diallel mating design in Mozambique. Results of this study showed that both GCA and SCA were significant for CBSD resistance. The Chigoma and Mulalei genotypes appeared as good candidates to be used as parental lines in breeding programs for CBSD resistance, while preserving other desired traits such as good yield and root characteristics[97].
To study the genetic control of resistance to CBSD, Kulembeka and colleagues evaluated cassava clones generated in a full diallel mating design with four parents at two locations (Chambezi and Naliendele) in Tanzania. The main finding of this study was that the GCA effects were more important than SCA in the genetic control of resistance to CBSD[53].
In Malawi, four parents with good flowering ability, low mean CBSD scores and good GCA for CBSD resistance were identified in 2012. They were either locally bred/improved (parents Phoso and Mkondezi) or introduced by the International Institute of Tropical Agriculture (IITA) (parents Silira and Mulola). The four parents were crossed, and the breeders selected 13 progenies with CBSD resistance and high storage root attributes for genetic advancement[98].
Dual infections of the viruses causing CMD and CBSD are commonly observed in cassava fields[99]. The only effective and sustainable way of controlling them is the development of dual resistant cassava cultivars[61]. In this regard, a CMD resistant parent (AR37-80) was crossed with a CBSD resistant parent (Namikonga) to generate F1 progenies with dual resistances. Crosses were carried out and evaluated for two seasons at the Tanzania Agricultural Research Institute (TARI) located at Naliendele (a CMD and CBSD hotspot) in Southern Tanzania. Majority of F1 progenies were CMD and CBSD resistant, and their resistance was not significantly different from their respective resistant parents. These progenies with dual resistance (Namar 050, Namar 110, Namar 200, Namar 334, Namar 371, and Namar 479), are important sources of resistance that could be further evaluated to determine farmer acceptance or exploited in future breeding programs[100]. The deployment of CBSD resistant cultivars have provided a significant contribution to cassava yield improvement in Uganda over the last decades. The evaluation of 32 cassava cultivars released from 1940 to 2019 in Uganda indicates an average annual genetic gain of 2.3% and 1.5% for CBSD foliar and CBSD root necrosis resistances respectively[101].
Inbreeding was used as another conventional breeding strategy against CBSD. Cassava is a cross pollinating plant which results in a high level of heterozygosity. It is known that inbreeding via consecutive self-pollination cycles eases the identification of useful recessive traits, either already present in the cassava gene pool or induced by mutagenesis[3,7,102]. This is achieved via (a) breaking down allelic combinations of individuals followed by a change in phenotypes; and (b) increasing the homozygosity of loci which eases the elimination of masked unwanted alleles[103] and, therefore, reducing the genetic load that was present in cassava genotype[3,7,102] . Despite the potential of selfing in cassava improvement, a severe inbreeding depression is the major challenge[104] arising from the accumulation of deleterious mutations in the genome[102,105] . Inbreeding depression drastically affects the survival of inbred genotypes in cassava[104] causing a shortage of breeding materials for the next selection phases. Nonetheless, research on inbreeding has shown some good potential. For instance, reportedly S1 clones have been reported to display good agronomic features including plant height, starch yield and dry matter content[104].
For cassava diseases, little but promising research has been carried out. For example, a study on inbreeding depression for severity caused by cassava leaf diseases revealed the predominance of additive genetic effects, highlighting the possibility of selecting transgressive individuals in S1 families[105]. Despite the amount of research that has been carried out on cassava inbreeding[104−107], very little has so far been achieved using this approach to enhance CBSD resistance. In Uganda, researchers assessed the possibility of exploiting inbreeding as a strategy for generating new sources of resistance to CBSD. A high proportion of generated seedlings was poor in terms of vigor, height and growth, and this was considered to be due to inbreeding depression and virus infection. Interestingly, within each family, a few S1 inbreds exhibited higher levels of resistance (based on foliar and root symptoms) to viral diseases[58]. The use of selfing in cassava remains an interesting approach to identify recessive genes for virus resistance as well as to carry higher breeding values in the parental material used for introgression of virus resistance[108].
In the last two decades, various studies were conducted on natural recessive resistance to plant potyviruses and this resistance was shown to be associated with polymorphism in the eukaryotic translation initiation factor genes (eIFs) which inhibits virus interactions with VPg, leading to the failure of the virus to infect the plant. The eIFs-mediated recessive resistance against potyviruses, closely related to ipomoviruses in the same family Potyviridae, was reported in several crops including pepper[109], lettuce[110] and tomato[111].
While conventional breeding have delivered important advances in CBSD resistance, there is a need to continue characterizing the novel sources of durable CBSD resistance by developing molecular markers to ease the introgression of CBSD resistance.
Regional strategies to mitigate cassava brown streak disease
-
Since the outbreak of CBSD in previously unaffected parts of East and Central Africa in the early 2000s, concerted regional efforts have been vital for managing the disease. For instance, selection of tolerant genotypes and use of open quarantine facility at Kibaha (Tanzania) was instrumental to readily provide tolerant varieties to be used as a short-term CBSD mitigation strategy in Tanzania and Mozambique[27].
Virus elimination was also used as a regional strategy to combat CBSD (Fig. 2a) and this elimination was implemented via the 5CP project (Cassava varieties and Clean seed to Combat CBSD and CMD project) targeting the eastern and southern African countries including Kenya, Tanzania, Uganda, Malawi and Mozambique. Using virus-indexing, tissue culture, chemotherapy and thermotherapy; 31 cassava varieties were successfully cleaned from virus infections. The virus elimination study was implemented in the UK (under controlled conditions) as a neutral venue and successfully cleaned plantlets were shipped back to the African partners for multiplication and subsequent distribution to farmers[43].
As part of the same project, a second batch of the 31 clones were cleaned up by the Kenya Plant Health Inspectorate Service (KEPHIS). Following successful virus elimination and indexing, about seventy-five in vitro plantlets per clone (from five countries) were sent to Genetic Technologies International Limited (GTIL), a private tissue culture lab in Kenya, and micro-propagated to produce ≥ 1,500 plantlets[112]. Successfully cleaned plants were shipped back to the partners in each of the five countries for hardening off and field multiplication in low disease pressure sites. Materials multiplied in this way were used to set up multi-locational trials for genotype by environment evaluations[38] and the top-performing clones in each country were advanced for national performance trials, official release, followed by distribution to farmers through seed systems. This study, therefore, contributed to the virus elimination, exchanging, release and dissemination of the best cassava germplasm at the regional level which contributed significantly to the management of both CMD and CBSD. In addition, the project built strong partnerships between breeders and virologists in national and international institutions that has ensured the successful exchange of elite germplasm. Combined with other breeding efforts implemented by NARS, initiatives of this kind will remain key approaches to mitigate CBSD in the region. Capacity building is a key component in these programs given that the breeding of virus resistance often requires a wide range of expertise including breeders, pathologists, entomologists and molecular biologists. Smallholder farmers pursuing mass production of planting material also require technical support and training on how they can maintain clean planting material for subsequent production. Both formal and informal seed multipliers should be trained to sustain the production of high-quality planting material in target production countries[113].
Genomic tools and NPBTs to advance the breeding for CBSD resistance
-
With conventional breeding, it generally takes 8−10 years to develop a new cassava variety[114] (Fig. 2c). Therefore, new breeding technologies appear as an attractive option to shorten the time needed to develop and release new cassava varieties (Fig. 2c)[115]. Technologies such as marker assisted selection, genomic selection, transgenesis, genome editing and others have been developed and applied to cassava. Several NPBTs have been deployed in order to develop CBSD resistant cassava varieties (Table 2).
Table 2. Summary of published studies on NPBTs towards CBSD management.
No Objective of the study Key findings Reference 1 To identify molecular markers associated with resistance against CBSD Two consistent QTLs were associated with CBSD root necrosis resistance on chromosomes II and XI, plus a putative QTL located on chromosome XVIII. [116] 2 To identify QTL associated with resistance to CBSD root necrosis and CBSD foliar symptoms Two QTLs were associated with only CBSD root necrosis (on chromosomes V and XII), seven QTLs were associated with only CBSD foliar symptoms (detected on chromosomes IV, VI, XVII and XVIII) and two QTLs that were associated with both CBSD foliar and root necrosis on chromosomes XI and XV. [117] 3 To map QTL associated with resistance to foliar and root necrosis CBSD symptoms Two QTLs (1 minor and 1 major) for foliar CBSD symptoms on chromosome XI and XVIII. The major QTL for foliar symptoms explained 12.87% of PHV at 6 MAP. [119] 4 To determine CBSD resistance-associated allele frequency and distribution and investigate the genetic relationships between some CBSD resistant genotypes A low allele frequency was putatively associated with Namikonga derived resistance to CBSD and there was a wide distribution of this frequency in the south-eastern and central African germplasm. [120] 5 To evaluate the potential of GS to enhance CBSD resistance via assessing the accuracy of 7 genomic prediction models Results showed good predictive ability of 0.40–0.42 for foliar CBSD severity and 0.31–0.42 for CBSD root severity at 6 MAP. [132] 6 To assess the use of genomic predictions of West African clones by using training data from a Ugandan population Higher genomic prediction accuracies for CBSD resistance were obtained by using optimized East African training populations. [133] 7 To assess the efficacy of GS for improving cassava for CBSD resistance and other traits Low predictive abilities for CBSD resistance (mean of 0.20 for
G-BLUP) were reported.[135] 8 To evaluate the association of 5 eIFs with cassava tolerance and susceptibility responses to CBSD Results showed that two SNPs in two genes were weakly associated with the CBSD but without any direct causal-effect relationship. [136] 9 To examine the power of diverse germplasm assembled
from two cassava breeding programs in Tanzania at
different breeding stages to predict traits and discover
quantitative trait loci (QTL)QTLs associated with CBSD resistance were found on chromosomes 9 and 11 while QTLs detected on chromosomes 2, 3, 8 and 10 were associated with resistance to CBSD root necrosis. Other three QTLs for dual resistance to CMD and CBSD were detected on chromosome 4 (one QTL) and chromosome 12 (2 QTLs). [118] 10 To generate eIF isoform (nCBP1,nCBP2 and nCBP1/nCBP2) mutants in line 60444 and assess their reponses upon challenge with CBSV Results showed that ncbp-1/ncbp-2 mutants exhibited not only delayed and attenuated CBSD aerial symptoms, but also reduced severity and incidence of storage root necrosis. [137] CBSD: Cassava brown streak disease; CBSV: cassava brown streak virus; QTL: Quantitative trait locus; GS: Genomic selection; MAP: Months after planting; SNP: Single nucleotide polymorphism; eIF: Eukaryotic translation initiation factors. Identification of quantitative trait loci (QTLs) associated with CBSD resistance
-
Studies on the identification of quantitative trait loci (QTL) associated with CBSD resistance were performed[116−119]. Those QTLs are inputs in marker-assisted breeding (MAB) as they are expected to ease the pyramiding of resistance QTLs into a sole genotype, thereby speeding up the breeding process for CBSD resistance[117].
It is within this context that two QTLs consistently associated with resistance against CBSD root necrosis on chromosomes 2 and 11, as well as a putative QTL that was located on chromosome 18 were identified[116]. After constructing a genetic map of QTLs for resistance to cassava diseases, two QTLs associated with only CBSD root necrosis resistance (on chromosomes 5 and 12), seven QTLs associated with only CBSD foliar symptoms resistance (on chromosomes 4, 6, 17 and 18) and two QTLs associated with both CBSD foliar and root necrosis resistance on chromosomes 11 and 15 were identified[117]. Training population was used to detect QTLs associated with CBSD resistance after phenotyping and genotyping. In fact, QTLs associated with CBSD resistance were found on chromosomes 9 and 11 while QTLs detected on chromosomes 2, 3, 8 and 10 were associated with resistance to CBSD root necrosis[118]. Two QTLs (one minor and one major) for foliar CBSD resistance on chromosomes 11 and 18 were detected. The major QTL for foliar symptoms explained 12.9% of phenotypic variation at 6 MAP. For root necrosis, all identified QTLs were considered minor as the phenotypic variance explained (PVE) was less than 10%[119]. A low allele frequency putatively associated with Namikonga-derived resistance to CBSD and a wide distribution of this frequency in the South-Eastern and Central (SEC) African germplasm has also been reported[120].
Several CBSD-resistance QTLs have so far been reported[116−119]. However, there is limited progress in integrating and deploying those QTLs through marker-assisted breeding programs. Because QTL applicability can be limited to specific genetic backgrounds and may be subjected to GxE interaction effects thereby limiting their genetic stability for selection, it is important to continue translational research to effectively convert QTL knowledge into practical breeding strategies[121−127].
Genomic selection
-
Genomic selection (GS) has been used in numerous plant breeding programs including cassava[128−130]. GS has been used to estimate the genomic prediction accuracies for different traits and reports showed that the prediction accuracies varied between measured traits. For instance, moderate to high genomic and pedigree accuracies (0.56–0.72 and 0.62–0.78) were reported for cassava dry yield and fresh root yield respectively[131]. In another study, low to moderate predictive abilities were reported for fresh root yield (0.4569 to 0.4756), dry root yield (0.4689 to 0.4818) and dry matter content (0.5655 to 0.5670)[115].
GS has been also used as a strategy to improve cassava virus resistance and promising results have been reported. While prediction accuracies have reached higher levels (0.26–0.40) for CMD[114] , reports on using GS for CBSD resistance showed low to high prediction accuracies. In this regard, using empirical data from 1301 cassava clones, the accuracy of seven genomic prediction models was assessed and a predictive ability of 0.31–0.42 for CBSD root severity was reported[132]. Also, a study conducted by Ozimati and colleagues showed higher genomic prediction accuracies for CBSD resistance using optimized East African training populations[133]. Optimization of the training population entails selecting breeding lines with phenotypic and genotypic data, that are representative of selected candidates in order to maximize the genomic selection[134]. In another study conducted in Uganda, low predictive abilities for CBSD resistance (mean of 0.20 for G-BLUP) were reported[135].
The findings in the above studies on GS show the potential of using this approach in cassava breeding albeit not all results were satisfactory. For CBSD resistance, low to high prediction accuracies were reported. Interesting results were obtained using optimized training populations, implying that cassava breeders will have to first optimize the training population in order to successfully exploit the full potential of GS for CBSD resistance. The immune resistance to CBSD found in training populations could be introgressed into susceptible cassava breeding clones through repeated backcrossings using the conventional method. During each cycle of the repeated backcrosses, a representative number of the F1 individuals should be backcrossed to different susceptible parents of heterozygous genetic backgrounds to limit inbreeding depression and select CBSD-resistant individuals with an adequate level of heterozygosity. Alternatively, genes conferring immune resistance can be isolated and stacked to susceptible parents through genetic transformation. With the advancement of New Genomic Techniques, the edition of CBSD susceptibility factors to generate new ideotypes without altering the genetic constitution of the crop could also be considered.
Eukaryotic translation initiation factor (eIFs)
-
Recessive resistance against viruses is broadly exploited in crops susceptible to potyviruses[138−140]. The eIFs genes are considered as key actors in plant potyvirus interactions as mutations in those genes can lead to total or partial resistance to potyviruses[141]. Useful mutations in eIFs genes have so far been reported in pepper, cucumber, lettuce, arabidopsis and tomato[109,110,142−144].
The eIF-based studies were performed to assess their association with CBSD resistance. The association of five eIF genes with cassava tolerance and susceptibility responses to CBSD was evaluated. Using non-synonymous SNPs, this study revealed no significant association between any SNP of the five eIF4E genes and the tolerance or susceptibility to CBSD. However, the study reported that two SNPs in two genes were weakly associated with the CBSD responses but did not have any direct causal-effect relationship[136]. Sheat and co-workers recently evaluated South-American genotypes with various levels of resistance. Those accessions need to be evaluated for variation in eIFs sequences in order to understand the basis of their resistance[54]. Though recessive resistance was reported in several crop species, it would be important to evaluate the potential of recessive resistance by selfing accessions carrying mutations in eIF sequences predicted to alter their interactions with VPgs.
Clustered regularly interspaced short palindromic repeats (CRISPR) - mediated genome-editing technology provides new opportunities for engineering plant disease resistance, and it is considered as one of the NPBTs that can speed up the introduction of desired traits into crops[145]. Several studies have been carried out using CRISPR genome editing technologies on cassava[137,146,147]. Gomez and colleagues used CRISPR/Cas9 to edit cassava eIF4E isoforms i.e novel cap-binding protein 1 and 2 (nCBP-1 and nCBP-2). This resulted in a reduction in CBSD symptom incidence and severity in the susceptible model cultivar 60444. The resulting nCBP-1/nCBP-2 mutants exhibited not only delayed and attenuated CBSD aerial symptoms, but also reduced severity and incidence of storage root necrosis[137].
Mutation breeding
-
Mutation breeding through gamma irradiation has been tested for variety development[148]. Most studies conducted on cassava mutation breeding have so far targeted yield and related traits[148−151]. Although mutation breeding has not been specifically applied as a tool for generating cassava virus resistance, it was notable that the primary product of two decades of mutation breeding research in Ghana, the cassava variety ‘Tek Bankye’, was highly susceptible to CMD[149]. Due to its random nature, an effective use of mutation breeding to generate virus resistant material will require screening large numbers of mutants in locations where the virus pressure is high or with cost-effective methods under greenhouse conditions. Importantly, identification and exploitation of recessive virus resistance will also require to self each mutant prior to exposure to viral pressure.
Other technologies for speeding up cassava breeding
-
Cassava is vegetatively propagated via stem cuttings[70,152]. This characteristic is advantageous for breeders because it permits to quickly get enough clonal materials to perform large scale crossings between selected parental lines. Breeders use crossing and seed propagation but getting desired crosses (traits) is not easy because of poor flowering, non-synchronization of flowering time and low pollen viability[152]. Moreover, the cassava life cycle is very long[114] and there is often a strong GXE component to flowering[74]. Methods that stimulate early flowering with an increased number of viable flowers could be instrumental to shorten the long cassava breeding cycle[153]. Given the long cassava breeding cycle, the so-called speed breeding techniques[154] hold a great potential to revolutionize cassava breeding.
One of the most important factors to consider for speed breeding in cassava is flower induction and boosting flower initiation in genotypes that are poorly flowering. Cassava flowering is associated with branching ability and growth type. However, erect growing genotypes are usually preferred over branching genotypes which further limit the potential to efficiently cross the parental material. Different approaches such as grafting, plant hormones, photoperiod extension[70,155] and genetic engineering such as introgression of flower-inducing genes in cassava[156,157] have been reported to induce cassava flowering, but not all of them are practically efficient. A useful technique that can be affordable and practical everywhere is the pruning of young cassava branches at the first flowering time[158].
Although not commonly used in cassava breeding programs, grafting was reported to enhance cassava flowering and promising results have been obtained as genotypes with profuse flowering habit can trigger flowering in low flowering genotypes through grafting[159,160]. Using red light emitting diodes (LEDs) and extended photoperiod conditions, it was possible to reduce the number of days to first branching in non or late flowering cassava genotypes[158]. The use of the flowering locus T (FT) was another strategy to induce early flowering in cassava. The over-expression of the Arabidopsis FT in transgenic cassava (model cultivar 60444) induced early flowering in glasshouse environment[156] as well as stimulation of branching, increased rate of flowering and increased viability of flowers[157]. The key role of the endogenous FT genes (MeFT1 and MeFT2) in flowering has also been confirmed using induction treatments such as extended photoperiod, cytokinin (BA) and silver thiosulfate (STS)[161]. Plant growth regulators (PGRs) were used to stimulate flowering in cassava. The anti-ethylene silver thiosulfate[162], cytokinin[163] and paclobutrazol[164] were reported to increase and sustain flower production in cassava. Notably, the optimum time for PGR application[162] and coupling PGR applications with pruning[163] are other important factors to consider for a successful stimulation of flowering in cassava. Most cassava breeding programs use a combination of EP, PGR and pruning, because other technologies appear less practical or constrained by regulation (i.e. transgenic expression of flowering genes).
Synchronization of flowering for parental genotypes is another key factor to consider in order to succeed and speed up cassava breeding. One strategy to synchronize flowering is the use of different planting dates. Genotypes that flower late should be planted ahead of time to match their flowering dates with early flowering genotypes[152]. This can only be successful if cassava breeders know the exact dates of flowering for the genotypes to be crossed. In addition, particular environments might favor flowering more than others[74], therefore optimal locations for flowering should be identified and used for flowering synchronization purposes. Poor flower synchronization can be mitigated through the use of extended photoperiod although it requires low temperature conditions which would only be applicable in high altitude environment or locations with seasonal winter[165].
Nowadays, pollen can be successfully stored in genebanks (cryopreservation in liquid nitrogen) without any damage to its genetic makeup[166]. To date, no protocol is available for cassava pollen cryopreservation, which could enable the crossing of genotypes with different flowering times. Cryopreservation can also facilitate the exchange of pollen materials between breeders working in different geographical locations[167]. Good quality and cryopreserved pollen grains would also be free of pathogens, thereby circumventing quarantine restrictions between countries.
-
CBSD remains an important constraint to cassava production in Eastern and Central Africa. Efforts to develop CBSD resistance will need to be an important part of modernized breeding approaches that aim to develop improved varieties with balanced trait sets that meet the needs of user-focused product profiles. This will require building regional capacity and provide long-term support to breeding programs, which should aim at providing millions of cassava farmers with varieties that combine high levels of CBSD resistance with optimal yield and market-preferred qualities. In order to achieve this ambitious goal, the following recommendations can be made:
(1) A network of African cassava breeders is required to share knowledge and experience about CBSD management across the continent. This network will also help breeders set collective strategies for CBSD management and create and/or enhance CBSD resistance in Africa.
(2) Access to advanced tools and technologies (such as sequencing facilities, genome-editing technologies, bioinformatics tools such as softwares, etc) by NARS and private companies will be paramount in mitigating CBSD and CMD impact as well as scaling up cassava improvement in Africa. These technologies should be adapted to circumvent the limitations posed by heterozygous progenitors.
(3) Building and/or strengthening the capacities of all stakeholders involved in cassava production (national and regional public and private institutions, seed multipliers, and farmers) will be crucial for cassava improvement in Africa. Investment by different stakeholders (governments, private sector, universities, international institutions, etc.) in cassava breeding is encouraged in order to develop cassava genotypes that are CBSD-resistant.
(4) Controlling the movement of planting materials is critical to prevent the introduction of CBSD-infected cuttings in a given country. This will require regular surveillance of each country’s border, observing strict quarantine measures and phytosanitary regulations throughout the distribution of cassava planting materials. Those measures should also set the scene for the development of a sustainable cassava seed system.
(5) Finally, the cassava community must address the issue of an efficient trait introgression in cassava breeding.
-
The authors confirm contribution to the paper as follows: study conception and design: Bizimana JP, Vanderschuren H; draft manuscript preparation: Bizimana JP, Nduwumuremyi A; Writing and editing: Bizimana JP, Vanderschuren H, Nduwumuremyi A, Ngapout Y, Shakir S, Kanju E, Nyirakanani C, Legg JP, Rukundo P, Shimelis H. All authors reviewed and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This study was funded by ARES (Académie de Recherche et d’Enseignement supérieur, Fédération Wallonie-Bruxelles, Belgium) via the project iCARE (Improved Cassava Virus Resistance-mitigation strategies and development of a disease-free seed system).
-
The authors declare that they have no conflict of interest.
-
Received 1 October 2023; Accepted 23 January 2024; Published online 26 February 2024
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Bizimana JP, Ngapout Y, Nyirakanani C, Shakir S, Kanju E, et al. 2024. Breeding strategies for mitigating cassava brown streak disease in Africa. Tropical Plants 3: e006 doi: 10.48130/tp-0024-0006
Breeding strategies for mitigating cassava brown streak disease in Africa
- Received: 01 October 2023
- Revised: 30 November 2023
- Accepted: 23 January 2024
- Published online: 26 February 2024
Abstract: Cassava (Manihot esculenta Crantz), is an important staple crop for about 800 million people worldwide, and a key commodity for the starch industry. However, the potential cassava production is limited by several biotic constraints amongst which cassava brown streak disease (CBSD) is the most economically important disease in Africa. To date, the most sustainable strategy to control CBSD is the use of resistant varieties and the supply of disease-free planting materials to cassava farmers. CBSD breeding activities were initiated in Tanzania in the 1930's and Manihot glaziovii species was used as a donor parent to generate resistant hybrids. Joint regional initiatives through variety development and virus-elimination methods have been vital for CBSD management in the last two decades. While conventional breeding is tedious and sources of broad-spectrum resistance are not always available to improve cassava, new plant breeding technologies (NPBTs) appears as a valuable and complementary tool to generate virus resistance as well as to speed up and scale up cassava breeding efforts towards CBSD resistance. This review presents the breeding strategies which have so far been used to manage the CBSD disease and discusses the advantages of integrating NPBTs in current cassava breeding programs to rapidly deliver CBSD-resistant varieties to farmers in Africa.
-
Key words:
- Cassava /
- CBSD /
- Breeding strategies /
- Conventional breeding /
- NPBTs












