-
Seed oils in plants are stored mainly as triacylglycerols (TAGs), which are formed by esterifying three fatty acids to glycerol-3-phosphate. They provide the energy needed for seed germination[1]. In recent years, the demand for plant oils has grown, making it important to study fatty acids and the transcription factors (TFs) that control fatty acid and TAG synthesis[2]. Among the TFs related to fatty acid regulation, WRINKLED1 (WRI1) and LEAFY COTYLEDON2 (LEC2) are the most widely studied and are regarded as master regulators[3,4]. Several other TFs are also reported to affect fatty acid synthesis during seed development[1,5]. However, the regulatory network remains incomplete, and the identity and roles of many TFs in fatty acid regulation are still unclear. It has been previously demonstrated that the overexpression of the seed master regulator LEAFY COTYLEDON2 (LEC2) in senescing Arabidopsis leaves led to triacylglycerol (TAG) accumulation and upregulation of downstream lipid biosynthetic genes. Transcriptome analysis of these LEC2-overexpressing plants revealed upregulation of 112 transcription factors, among which AtHAT2 expression increased by 1.4-fold (log2 scale)[4]. In Arabidopsis thaliana, a total of 48 homeodomain-leucine zipper (HD-Zip) genes have been identified and are classified into four major subfamilies: HD-Zip I through IV[6]. Each family is further divided into subgroups. Notably, AtHAT2 was first identified as an auxin-inducible gene[7], which belongs to the HD-Zip IIγ, and its expression is further enhanced under shade conditions through the auxin signaling pathway[8]. While the HD-Zip II family is known to participate in auxin-mediated developmental processes and the shade avoidance response[9], its role in seed development has not been explored, suggesting a potential role for AtHAT2 in seed lipid metabolism.
The amino acid sequence encoded by At5g47370 (AtHAT2) was analyzed and found to comprise 283 residues. Domain analysis using UniProt (P46601-1) identified a repression-related EAR motif (positions 10–14), a homeobox DNA-binding domain (127–186), a leucine zipper dimerization domain (194–215), and a conserved CPSCE motif (Cys-Pro-Ser-Cys-Glu), the latter being involved in redox sensing[10] (Fig. 1a). In Arabidopsis protoplasts, GFP expressed under the control of the CaMV 35S promoter was distributed throughout the cytoplasm. In contrast, the AtHAT2–GFP fusion protein, driven by the same promoter, displayed fluorescence signals that overlapped with the nuclear RFP marker (Fig. 1b), indicating that AtHAT2 is targeted to the nucleus, consistent with its role as a transcription factor.
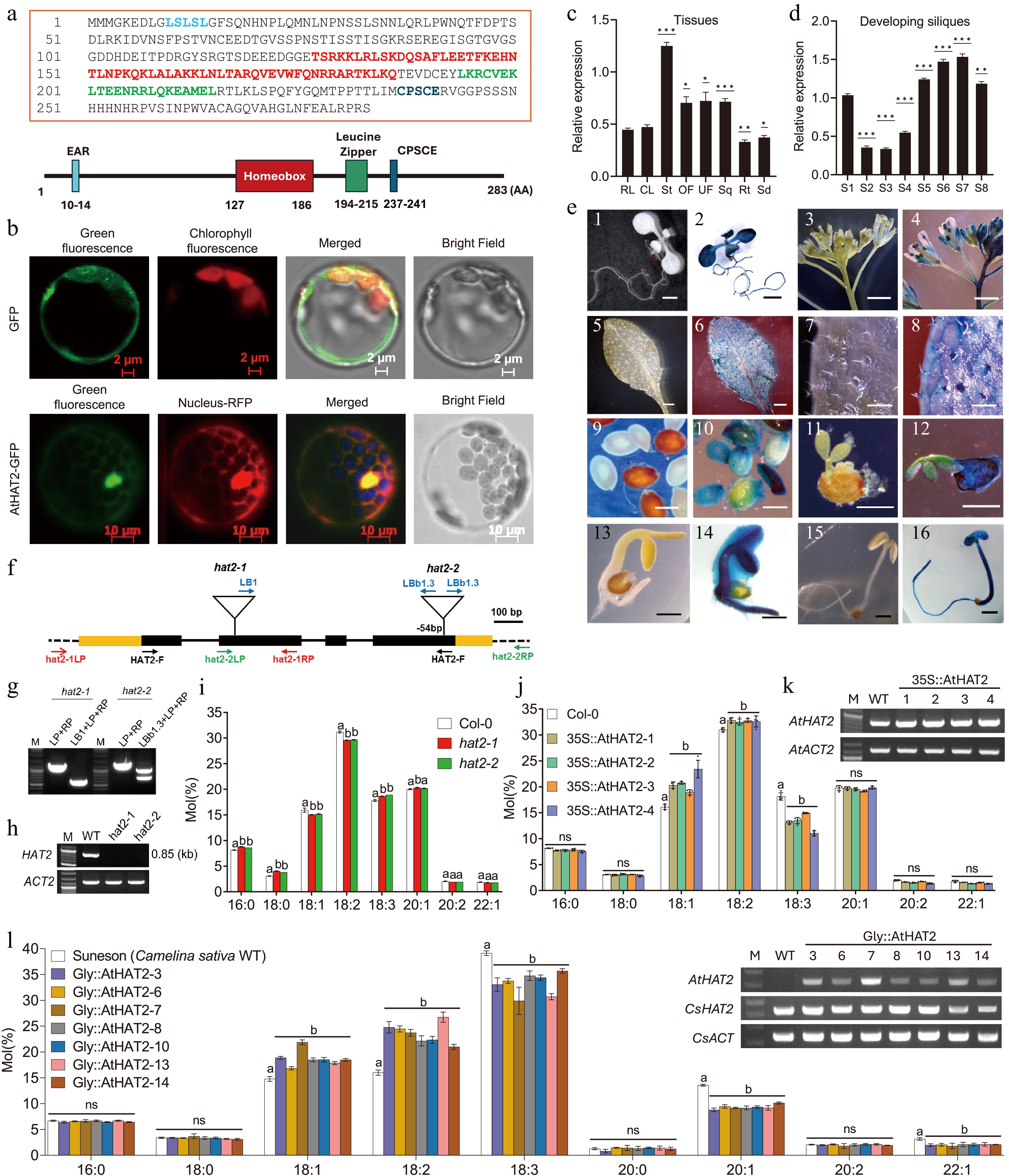
Figure 1.
Molecular and functional characterization of the AtHAT2 gene and its role in seed fatty acid composition.
(a) Amino acid sequence and schematic representation of the AtHAT2 protein. Conserved motifs are highlighted: EAR motif (blue), Homeobox domain (red), Leucine zipper (green), and CPSCE motif (navy). (b) Subcellular localization of GFP protein (control) and AtHAT2-GFP fusion protein (AtHAT2 positive) in Arabidopsis protoplasts. Green: GFP, AtHAT2-GFP; Red: chlorophyll auto-fluorescent, nuclear marker NLS-RFP. Scale bar = 2 μm (control); 10 μm (AtHAT2). (c) RT-qPCR analysis of AtHAT2 expression in various tissues: RL, rosette leaf; CL, cauline leaf; St, stem; OF, open flower; UF, unopened flower; Sq, silique; Rt, root; Sd, seedling. Statistical significance was determined by a sample t-test using the RL sample value as the control. (d) Temporal expression pattern of AtHAT2 during seed development. Statistical significance was determined by a sample t-test using the S1 sample value as control. (e) Histochemical GUS staining in AtHAT2::GUS transgenic and wild-type Arabidopsis plants. Tissues: (1), (2) seedling; (3), (4) inflorescence; (5), (6) rosette leaf; (7), (8) trichome; (9), (10) dry seed; (11), (12) seed coat and embryo; (13), (14) 3-day-old dark-grown seedlings; (15), (16) 5-day-old dark-grown seedlings. Scale bars = (1), (2): 0.25 mm; (3)–(6), (15), (16): 1 mm; (7)–(14): 0.5 mm. (f) Gene structure of hat2-1 and hat2-2 T-DNA insertion mutants. Exons (black boxes), introns (yellow boxes), T-DNA insertion sites with respective left borders (LB, LBb1, LBb1.3), and gene-specific primers (hat2-1LP/RP, hat2-2LP/RP) are indicated. (g) Genotyping of hat2 mutants using allele-specific primers. (h) RT-PCR analysis of AtHAT2 expression in wild-type and homozygous hat2 mutants. (i) Fatty acid composition of seeds from wild-type (Col-0) and hat2 T-DNA mutants. (j) Fatty acid composition of seeds from Col-0 and 35S::AtHAT2 overexpressing Arabidopsis lines. (k) RT-PCR analysis of AtHAT2 expression in 35S::AtHAT2 transgenic lines. Line numbers indicate individual transgenic events. AtACT2, the Arabidopsis thaliana ACTIN gene, was used as a control. (l) Fatty acid composition of seeds from wild-type Camelina sativa (cv. Suneson) and AtHAT2 seed-specific overexpression lines (Gly::AtHAT2). (M) RT-PCR analysis of AtHAT2 expression in Gly::AtHAT2 transgenic lines. Line numbers indicate individual transgenic events. CsACT, Camelina sativa ACTIN gene, was used as a control. All data are presented as mean ± SD (n = 3–5 biological replicates). Statistical significance was determined by two-way ANOVA followed by Dunnett’s multiple comparison test (p = 0.05). Groups not sharing the same letter are significantly different; for example, groups labeled 'a' and 'b' differ significantly, while 'a' and 'ab' do not.RT-qPCR expression profiling revealed that AtHAT2 is expressed in diverse tissues, including rosette and cauline leaves, roots, seedlings, flowers, siliques, and stems, with the highest levels detected in stems (Fig. 1c). Notably, AtHAT2 expression was elevated in both vegetative and reproductive tissues, particularly in stems, flowers, and developing siliques, suggesting a role in seed development. To examine this in greater detail, AtHAT2 expression was analyzed across eight silique developmental stages. Expression was dynamic, showing an initial peak at early embryogenesis (stage S1), a decline through stages S2–S4, and a subsequent increase from stage S5, reaching a maximum at stage S7 (Fig. 1d). Promoter–GUS fusion analysis further confirmed AtHAT2 expression in multiple tissues, including young leaves, roots, trichomes, veins, pistils, stamens, and developing embryos (Fig. 1e). Together, these results indicate that AtHAT2 has broad expressions and plays a significant role in regulating seed development.
To investigate the functional role of AtHAT2, two homozygous T-DNA insertion mutants, hat2-1 and hat2-2, with insertions in exons 2 and 4, respectively, were identified and confirmed via PCR and RT-PCR (Fig. 1f–h). Fatty acid profiles of mature seeds from hat2 mutants were analyzed by gas chromatography. Compared to wild-type (WT), hat2-1 and hat2-2 mutants showed slight reductions in 18:1 and 18:2 fatty acids and significant increases in 16:0, 18:0, 18:3, and 20:1 fatty acids (Fig. 1i, Supplementary Table S1).
Four independent Arabidopsis transgenic lines overexpressing AtHAT2 under the CaMV 35S promoter (35S::AtHAT2) were generated. RT-PCR analysis confirmed higher AtHAT2 transcript levels in these lines compared with the WT (Fig. 1k). Fatty acid profiling revealed that, unlike the hat2 knockout mutant, the 35S::AtHAT2 lines exhibited increased proportions of 18:1 and 18:2 fatty acids and reduced levels of 18:3 fatty acids relative to the WT (Fig. 1j, Supplementary Table S2). For example, 18:1 content rose from 16.1% in WT to 19.0%–23.4% in transgenic lines, while 18:3 decreased from 18.2% to 11.1%–15.0%. These changes were opposite to those observed in hat2 mutants, suggesting that AtHAT2 modulates fatty acid composition in seeds.
To test the potential application of AtHAT2 in oil crops, AtHAT2 was placed under the control of the seed-specific glycinin promoter and introduced into Camelina sativa (Gly::AtHAT2). Red fluorescence from T1 seeds allowed visual selection of transgenic lines, which were further confirmed by RT-PCR (Fig. 1m). GC analysis of T2 seeds from seven independent lines showed elevated levels of 18:1 (16.9%–21.9%) and 18:2 (21.0%–26.7%) compared to the WT (14.8% and 16.0%, respectively), and a concurrent reduction in 18:3 content (29.9%–35.7% vs 39.1% in WT) (Fig. 1l, Supplementary Table S3).
Although Arabidopsis and Camelina share the same fatty acid species, they differ in the relative proportions. Camelina generally has lower 18:2 and higher 18:3 and 20:1 than Arabidopsis. Nonetheless, seed-specific overexpression of AtHAT2 in Camelina recapitulated the effects seen in Arabidopsis, with increased 18:1 and 18:2 and decreased 18:3 and 20:1. Together, these results demonstrate that AtHAT2 regulates seed fatty acid composition in both Arabidopsis and Camelina, promoting accumulation of 18:1 and 18:2 while reducing 18:3 and 20:1.
To investigate the molecular basis of altered fatty acid composition in hat2 mutants and 35S::AtHAT2 overexpression lines, microarray analysis was conducted using RNA from developing siliques of wild-type, hat2-1, hat2-2, and 35S::AtHAT2 plants. Comparative transcriptome profiling revealed no significant differences in the expression of genes directly involved in fatty acid and triacylglycerol (TAG) biosynthesis (Supplementary Table S4). In particular, the transcript levels of FAD2, FAD3, and FAE1 − which catalyze the conversion of 18:2 to 18:3 and the elongation of 18:1 to 20:1 − were unchanged across all genotypes (Supplementary Fig. S1).
These findings indicate that the altered proportions of 18:1, 18:2, 18:3, and 20:1 in AtHAT2 mutants and overexpression lines are not due to direct transcriptional regulation of fatty acid metabolic genes. Instead, AtHAT2 likely affects seed lipid composition through indirect mechanisms, such as developmental regulation or upstream metabolic adjustments that influence substrate availability for desaturation and elongation. Taken together, our results suggest that AtHAT2 functions as a novel regulator of fatty acid homeostasis, acting independently of the canonical transcriptional control of lipid biosynthetic enzymes. The underlying mechanisms remain to be clarified and warrant further investigation.
HTML
This work was supported by the National Research Foundation of Korea (NRF) through the Mid-Career Researcher Program (RS-2025-00514459), and the Basic Research Laboratory Program (RS-2024-00410854), as well as by the Rural Development Administration of the Republic of Korea through the New Breeding Technologies Development Program (RS-2024-00322277).
-
The authors confirm their contributions to the paper as follows: project conception and research plans: Kim HU; experiments conducted: Kim WN, Park M-E, Kim HU; draft manuscript preparation: Kim WN, Park M-E; manuscript review and editing: Kim HU. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files.
-
The authors declare that they have no conflict of interest.
-
#Authors contributed equally: Won Nyeong Kim, Mid-Eum Park
- Supplementary Table S1 Fatty acid composition in mature seeds of hat2-1 and hat2-2 Arabidopsis T-DNA insertion mutants.
- Supplementary Table S2 Fatty acid composition in mature seeds of Arabidopsis transgenic lines overexpressing HAT2 (35S::AtHAT2).
- Supplementary Table S3 Fatty acid composition in mature seeds of Camelina transgenic lines overexpressing AtHAT2 under the seed-specific glycinin promoter (Gly::AtHAT2).
- Supplementary Table S4 Microarray analysis of developing siliques from wild type, hat2-1, hat2-2, and 35S::AtHAT2 lines.
- Supplementary Fig. S1 Relative expression analysis of FAD2, FAD3, and FAE1 in wild type, hat2-1, hat2- 2, and 35S::AtHAT2 lines. Expression levels were derived from microarray analysis. No statistically significant differences were detected among the four genotypes.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press on behalf of Yunnan Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Kim WN, Park ME, Kim HU. 2025. The HD-Zip II transcription factor AtHAT2 modulates seed fatty acid composition in Arabidopsis and Camelina. Agrobiodiversity 2(3): 59−61 doi: 10.48130/abd-0025-0008 |













