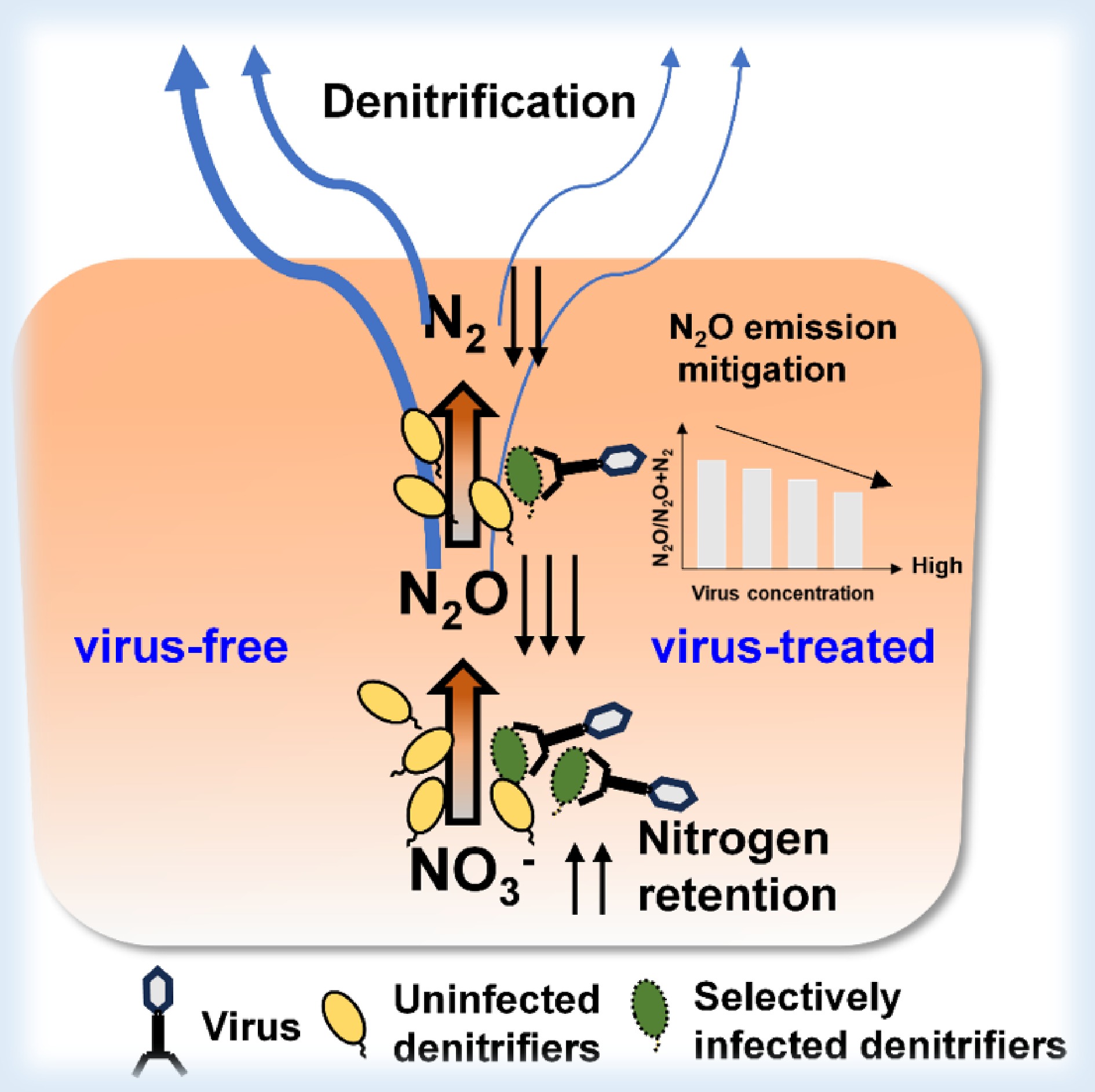
-
Denitrification, the stepwise reduction of nitrate (NO3−) to nitrite (NO2−), nitrous oxide (N2O), and finally dinitrogen (N2), plays a fundamental role in soil nitrogen losses and the emission of N2O, a potent greenhouse gas[1]. Viruses, as predators of microbes, are abundant in soils and potentially influence denitrification through the following pathways. (1) Infection and lysis: Viruses may infect denitrifying microbes, causing lysis of the host cells, which shapes the composition of the denitrifying community and alters the expression of functional genes[2]. (2) Horizontal gene transfer: During infection, viruses may introduce auxiliary metabolic genes (AMGs) into the host cells, affecting the functional gene expression of infected denitrifying microbes[3,4]. (3) Release of intracellular carbon and nitrogen: Widespread viral infection and microbial lysis can release dissolved organic carbon and nitrogen from microbial cells, providing substrates for denitrifiers, thereby influencing N2O and N2 emissions[5−7].
Current research on the microbial mechanisms underlying soil denitrification and N2O emissions primarily focus on the community structure and functional gene expression of denitrifying bacteria and fungi[8]. Although viruses are known to be abundant in the soil, their role in soil nitrogen cycling, including denitrification, remains poorly understood. Previous studies have employed virus inoculation methods to investigate the effects of viruses on greenhouse gas emissions[7]. To date, no controlled studies have examined the role and mechanisms by which viruses influence soil denitrification and the composition of its gaseous products. Addressing this knowledge gap is critical for advancing our understanding of microbial regulatory mechanisms in denitrification and developing strategies to mitigate soil nitrogen losses and N2O emissions.
To fill this knowledge gap, this study designed a control experiment with four levels of viral abundance. Using direct N2 measurements and microbial genomic analysis, we aimed to quantify the effects of viruses on N2O and N2 emissions, product ratios (N2O:N2O+N2), and the underlying mechanisms.
The soil used for the experiments was collected from the Luancheng Agro-Ecosystem Experimental Station (37°90′ N, 114°70′ E) in the North China Plain, a typical grain production region characterized by high nitrogen fertilizer application (400 kg N ha−1·yr−1), significant nitrogen loss, and high N2O emissions. Viruses were extracted from 200-g soil samples using a saline magnesium (SM) buffer[6], with a soil-to-buffer ratio of 1:1. The soil–buffer mixture was shaken for 2 h, followed by centrifugation and filtration through 0.22-µm membranes to separate the viral particles. The viral extract was concentrated using a 100-kDa Amicon centrifugal filter[9]. The concentrated viral solutions were divided into two groups: sterilized and nonsterilized. The sterilized group was autoclaved at high temperature and pressure, a widely used method for viral sterilization[6].
For each treatment, 10 g of fresh soil was placed into anaerobic bottles, and each sample received 10 mL of the viral solution to achieve the desired concentrations: 100% (10 mL live virus extract), 50% (5 mL live and 5 mL inactivated virus extract), 25% (2.5 mL live and 7.5 mL inactivated virus extract), 12.5% (1.25 mL live and 8.75 mL inactivated virus extract), and the control (CK) (10 mL inactivated virus extract). It is important to note that the virus concentrations used in this study are higher than typical natural field-level viral abundances. The highest viral concentration in the treatment was double that of the control, but it did not reach the upper limits of natural field concentrations. The concentrations used in our experiment represent an increase over baseline levels rather than extreme or unrealistic viral abundances, which could provide more insight into the effects of adding viruses under experimental conditions.
All bottles were sealed, and the headspace was flushed with helium (99.99%) to create anaerobic conditions. Each treatment had three replicates. The bottles were incubated at 25 °C for 48 h, with periodic mixing to ensure uniform conditions. During the incubation period, N2O and N2 emissions were measured every 3 h using an automated robotic system with an Agilent 7890A gas chromatograph, coupled with both an electron capture detector (ECD) and a thermal conductivity detector (TCD). The column temperature was set to 50 °C, and the carrier gas was 99.99% helium[10]. Denitrification product ratios (N2O:N2O+N2) were calculated to assess the efficiency of the denitrification process.
After incubation for 15 days, total DNA was extracted from 0.5 g of soil using a DNA extraction kit, and the DNA was sent to Magigene for metagenomic sequencing with a sequencing depth of 30 Gb. The metagenomic data analysis methods are provided in the Supplementary File 1.
Data were analyzed using one-way analysis of variance (ANOVA) to compare the effects of viral abundance on N2O and N2 emissions, product ratios (N2O : N2O+N2), and microbial community structure.
Our results show that the addition of viruses significantly reduced soil N2O and N2 emissions, and altered the product ratios (N2O:N2O+N2) (Fig. 1a–c). The 100% viral treatment reduced cumulative N2O emissions by 20% compared with the control. (Fig. 1a). At the end of the incubation, soil NO3− content was higher in the virus-treated soils compared with the control (Supplementary Fig. S1). This result indicates that viruses can regulate soil denitrification processes and their products' composition, thereby contributing positively to soil nitrogen retention and greenhouse gas mitigation.
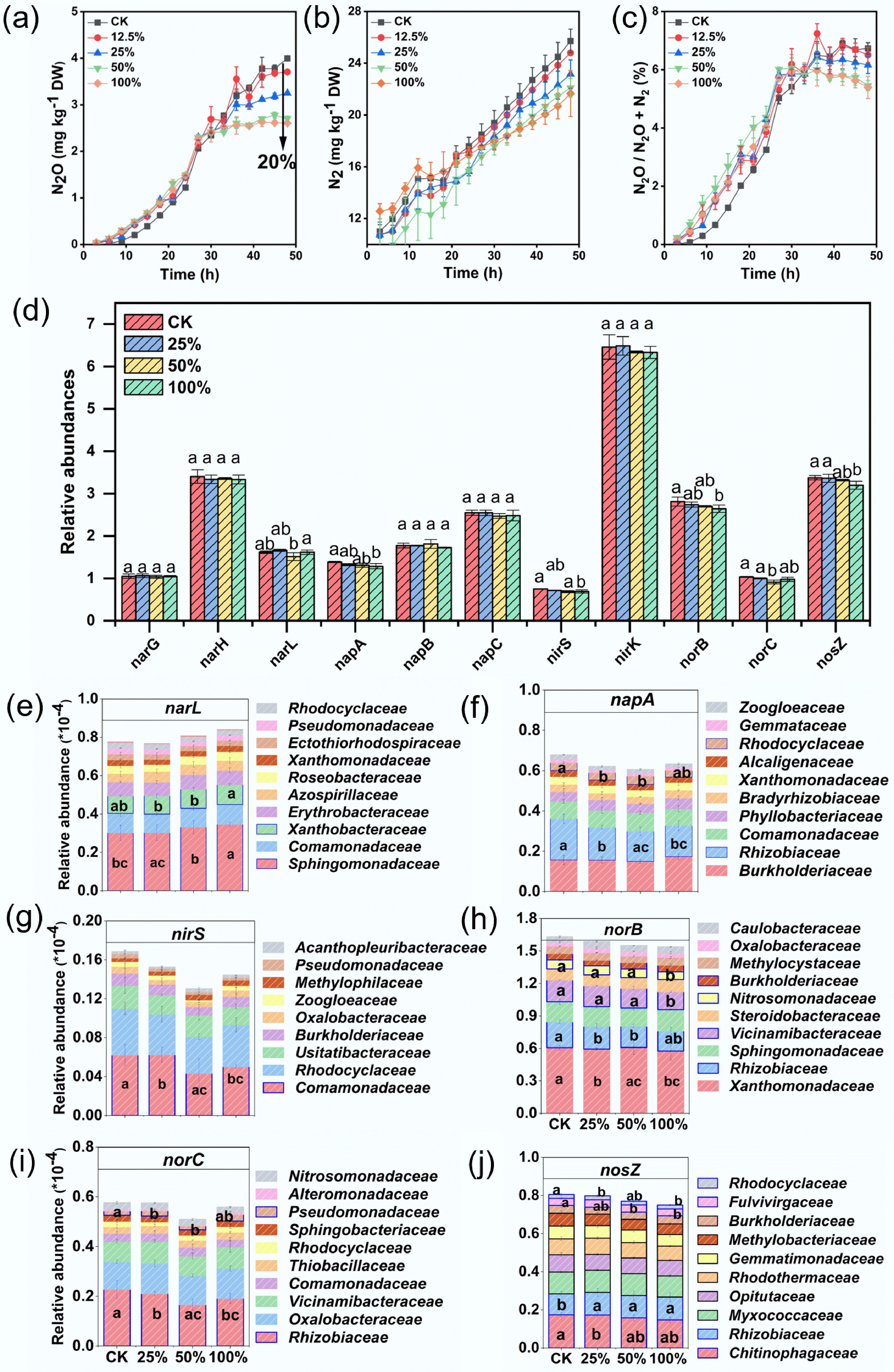
Figure 1.
Effects of viral concentrations on the end products of denitrification, functional gene abundance, and denitrifying community composition. Viral treatments are represented as percentages: 100% indicates 10 mL live virus extract + 0 mL inactivated virus extract (similar definitions apply for other percentages). (a–c) Gaseous products of denitrification and their ratios. (d) Changes in the abundance of key denitrification-related functional genes under varying viral treatments. (e–j) Relative abundance of microbial taxa harboring denitrification-related genes under different viral treatments. Different letters above the data points indicate significant differences at p < 0.05.
In theory, the effects of viruses on microbial functions are mediated through three mechanisms. One mechanism is the release of soluble carbon and nitrogen from the host cells' contents through viral lysis, which subsequently influences the denitrification process[5,6,11]. In this study, there were no significant differences in the soil's dissolved organic carbon (DOC) content across treatments, indicating that the organic matter released through viral lysis was insufficient to significantly affect soil DOC levels (Supplementary Fig. S1). Thus, the impact of viruses on denitrification is unlikely to be driven by the release of soluble carbon and nitrogen from the host cells. Another potential pathway for viruses' influence on denitrification is the transfer of AMGs from viruses to the host cells during infection, enhancing specific metabolic functions, such as denitrification, in the host[3]. However, no denitrification-related viral AMGs were detected in this study (data not shown). This absence may be attributed to the lower significance of denitrification processes for viral nutrient acquisition compared with metabolic pathways related to carbon and phosphorus cycling[3,12]. As such, denitrification-related AMGs may be less abundant and exert limited effects on host cells.
The most direct effect of viruses on the hosts' denitrification functionality is through selective infection of key denitrifying microbial taxa, thereby restructuring the community and altering functional gene expression[13]. Our metagenomic analysis supports this mechanism, revealing that increasing viral abundance significantly shifted the community composition (Fig. 2, Supplementary Figs S2 and S3), with notable declines in major denitrifying families such as Sphingomonadaceae, Xanthobacteraceae, Rhizobiaceae, and Rhodocyclaceae (Fig. 1e, f). Moreover, viral inoculation significantly increased the abundance of Megaviricetes and Pokkesviricetes (Fig. 2a), which are predicted to infect denitrifiers, according to host-linking analysis[14,15]. Correspondingly, the abundance of Pseudomonadota declined significantly (p < 0.05; Fig. 2b), consistent with reductions in the abundance of nirS and norB genes (Fig. 1e, f). This suggests that viral lysis of Pseudomonadota, linked to nirS and norB, may suppress early denitrification. These findings indicate that viruses reduce denitrification by selectively targeting key functional microbes. However, potential off-target effects on other functional microbial groups, such as nitrogen-fixing bacteria, were not assessed and warrant further investigation.
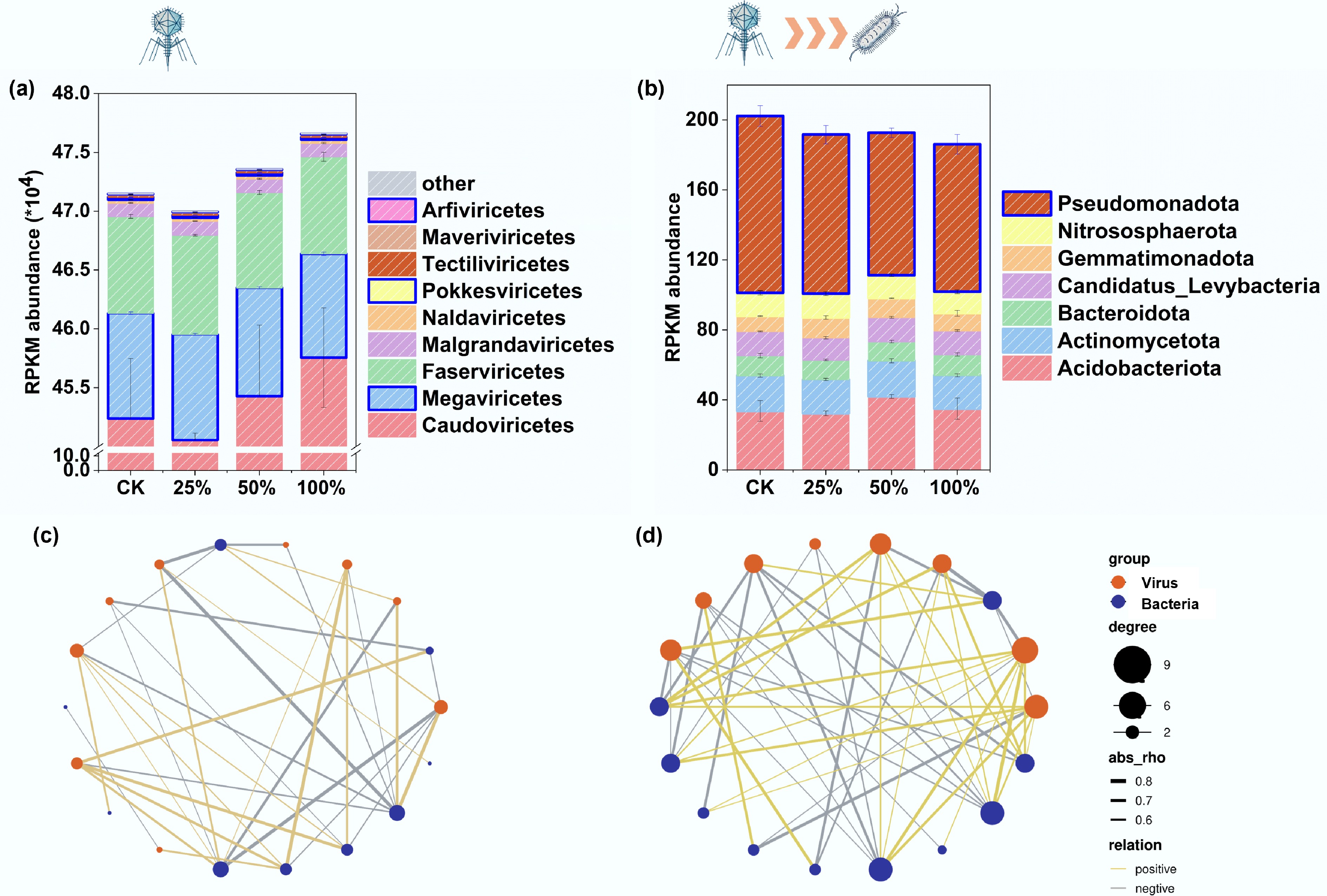
Figure 2.
Characterization of viral and host communities and virus–microbe interaction networks under varying viral concentrations. (a) Viral community composition, (b) hosts of Megaviricetes, (c) virus–microbe networks at 0% and 25% viral concentrations, and (d) virus–microbe networks at 50% and 100% viral concentrations. Different colors denote significant differences (p < 0.05) among treatments.
Network analysis further revealed that high viral treatments resulted in more complex virus–microbe interaction networks compared with the low viral and control treatments (Fig. 2c, d). Specifically, the number of network nodes increased from 70 in the control to 102 in the high viral treatment, representing a 46% increase and indicating more extensive virus–host associations. This enhanced complexity is likely driven by viral infection dynamics rather than auxiliary metabolic functions, as viruses associated with denitrification-related processes were predominantly smaller than 10 kb and generally lacked virus-like AMGs.
This study underscores the previously underappreciated role of viruses in mitigating soil nitrogen losses and N2O emissions during denitrification. Our findings highlight the necessity of incorporating viral dynamics into soil biogeochemical models to enhance the accuracy of predictions related to nitrogen losses and greenhouse gas emissions. Furthermore, the application of phage therapy emerges as a promising strategy for mitigating N2O emissions and nitrogen losses from agricultural soils. Extrapolating the observed 20% N2O reduction to cereal croplands of China (annual N2O–N emissions estimated at 1.0−1.5 Tg)[7] yields a theoretical mitigation potential of 0.2−0.3 Tg N2O–N yr−1, highlighting the potential large-scale climate benefits of virus-based regulation if it is proved to be effective under field conditions. Enhancing the feasibility of this approach may require stabilizing viral particles through encapsulation in clay minerals[9] and formulating phage cocktails that target high-emission microbial taxa to improve specificity and cost-efficiency. Given that this study only controlled the temperature and anaerobic conditions, future work should assess whether viral regulation remains effective under more variable soil environments (e.g., pH and moisture) and fertilization regimes across land-use types.
HTML
-
Accompanies this paper at: https://doi.org/10.48130/nc-0025-0002.
-
The authors confirm their contributions to the paper as follows: study conception and design: Song W, Yao J, Qin S; data collection: Song W, Yao J, Fu Y; analysis and interpretation of results: Song W, Yao J; draft manuscript preparation: Song W. All authors reviewed the results and approved the final version of the manuscript.
-
The data that support the findings of this study are available on request from the corresponding author.
-
This work was supported by the National Natural Science Foundation of China (42277360), the National Natural Science Foundation of Hebei Province (D2022503014), China Postdoctoral Science Foundation (2024M763523), and Hebei Province Postdoctoral Science Foundation (B2024005039).
-
The authors declare that they have no conflict of interest.
-
Viral inoculation reduced denitrification N loss and N2O emissions by up to 20%.
Viral inoculation shifted microbial community structure, enriching Megaviricetes and Pokkesviricetes.
Viruses selectively infected denitrifying microbes Pseudomonadota, reducing N2O formation.
Phage therapy could offer a promising approach to reducing soil N2O emissions.
-
Full list of author information is available at the end of the article.
- Supplementary File 1 Metagenome and virome analysis.
- Supplementary Fig. S1 Concentrations of soil nitrate (NO3−), DOC, pH and SOC at the end of the incubation. Data represent the mean ± SD (n = 3). Different letters above data points indicate significant differences at p < 0.05.
- Supplementary Fig. S2 NMDS (Non-metric Multidimensional Scaling) analysis of viral community composition across different viral concentration treatments.
- Supplementary Fig. S3 Microbial community composition at the Class level across different treatments. Each bar represents the relative abundance of microbial classes within each sample, distinguished by distinct colors. Data are presented as mean ± standard deviation (n = 3).
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Song W, Yao J, Fu Y, Qin S. 2025. Viruses mitigate soil nitrogen loss and N2O emissions during denitrification by selectively infecting denitrifiers. Nitrogen Cycling 1: e004 doi: 10.48130/nc-0025-0002 |