-
Hypocotyl elongation drives seedling emergence, light capture, and structural integrity in plants, involving coordinated cellular expansion[1], cell wall modification[2,3], and microtubule dynamics[4]. It is regulated by diverse endogenous hormonal and environmental stimuli through a central circuit of interacting transcriptional regulators[5]. In eudicot seedlings, the hypocotyl connects the cotyledons to the roots, serving as a vital conduit for transport and constituting a key agronomic trait that influences plant stature and lodging resistance[6].
Brassinosteroids (BRs), a class of polyhydroxylated steroidal hormones, regulate cell elongation and division[7−12]. When the plasma membrane (PM)-localized receptor kinase BRASSINOSTEROID INSENSITIVE1 (BRI1) senses BRs, the signal is transmitted stepwise to a plant-specific transcription factor, BRASSINAZOLE-RESISTANT1 (BRZ1) and BRI1-EMS-SUPPRESSOR 1 (BES1/BZR2)[13−15]. BR-induced BZR1/BES1 dephosphorylation targets thousands of downstream genes to control various physiological processes in plants[16,17]. BZR1/BES1 directly upregulates EXPANSINS (EXPs), which encode cell wall proteins involved in plant cell extension[18]. Moreover, the BR-mediated BES1–cell elongation-related protein (CERP)–EXPA3 signaling cascade controls plant cell elongation in cotton and Arabidopsis[19].
Emerging evidence suggests brassinosteroid pathways could potentially modulate hypocotyl elongation in Brassica rapa (B. rapa) subspecies. The Brassicaceae family possess rich germplasm resources and exhibit diverse morphological variations, including leafy vegetables and root vegetables, which are widely consumed[20−22]. In Brassica crops, excessive hypocotyl elongation compromises plant lodging resistance, whereas insufficient elongation hinders mechanized harvesting. Consequently, hypocotyl length is a critical agronomic trait affecting harvesting efficiency, particularly during mechanical cutting at the hypocotyl or lower stem regions, and has been a longstanding focus for breeders[23]. To date, the studies on hypocotyl elongation in B. rapa show that light, temperature, water potential, auxin, BR, and cell wall dynamics affect hypocotyl elongation[24−28].
Turnip (B. rapa ssp. rapa) and Chinese cabbage (B. rapa ssp. pekinensis), both of which belong to the Brassica genus, have big interspecies differences in hypocotyl length; thus, they are suitable for the exploration of the key genes involved in interspecies differences in hypocotyl regulation. In this study, it was found that BR signaling is associated with the contrasting hypocotyl length phenotypes of these two species. To explore the genetic basis of BR signaling in turnip and Chinese cabbage, and the differential control of hypocotyl elongation, BR biosynthesis and signaling pathways between these species were systematically compared at the transcriptional, physiological, cytological, and molecular levels. The present results showed that, compared with that in Chinese cabbage, BR signaling is more effectively transduced in turnip and promotes hypocotyl elongation.
-
An inbred turnip line (MM) with a long hypocotyl and an inbred Chinese cabbage line (BY) with a short hypocotyl were selected for studying hypocotyl elongation. These lines grown in soil were germinated at 25 °C and planted under LD (16 h light, 25 °C/8 h dark, 18 °C) conditions. Plants grown on ½ MS culture medium were washed with ddH2O three times after being disinfected with 10% NaClO for five minutes, and were then placed under LD conditions.
Hypocotyl length measurements
-
For plants growing in soil, the hypocotyl length was directly measured with a vernier caliper at 3, 8, and 15 days after sowing (DAS). Plants grown on ½ MS culture medium were photographed at 10 DAS, and the hypocotyl length was measured with ImageJ software. Transgenic seedlings were grown on ½ MS medium containing kanamycin for the first 5 d. Thereafter, the remaining green seedlings were transferred to standard MS medium, and grown for another 2 d under long-day conditions.
Cell observation
-
MM and BY seedlings were grown in soil for visualization on paraffin slides. Samples of fresh hypocotyl tissue were immediately put into a fixative solution at 3, 8, and 15 DAS. The subsequent experiments were carried out by Wuhan Servicebio Technology Co., Ltd (Wuhan, China).
Plants were cultured on ½ MS medium for 10 d for the compound (BL and Brz) treatment experiments. The hypocotyl epidermis was torn and photographed under transmitted light via microscopy.
BL and Brz treatments
-
Stock solutions of BL and Brz were prepared in 80% ethanol and DMSO, respectively. The corresponding concentrations of BL or Brz were added to ½ MS culture medium for treatment. The concentrations of BL were 0, 1, 10, 100, and 1000 nM, with or without supplementation of 1 μM Brz. The concentrations of Brz used were 0, 0.1, 1, 2, and 10 μM. Seeds were neatly stacked on agar plates supplemented with the abovementioned concentrations of BR and Brz for 10 d and were then photographed to measure the hypocotyl length.
Quantification of BRs in plants
-
Hypocotyl samples from MM and BY plants grown under LD conditions were collected at 6 and 15 DAS, with three biological replicates per time point. BRs were extracted from approximately 100 mg of ground hypocotyl tissue with a cold methanol : acetonitrile (50:50, v/v) solution. The samples were collected and rapidly frozen in liquid nitrogen. LC‒MS analysis and quantification of plant hormones were performed by Wuhan Metware Biotechnology Co., Ltd (Wuhan, China).
Transcriptome analysis
-
Two inbred lines, MM (turnip) and BY (Chinese cabbage), were germinated at 25 °C and planted in soil for growth under LD conditions. The hypocotyls of these plants were collected in the morning at 3 (fast elongation stage), 8 (elongation stage), and 15 DAS (ending stage) for transcriptome analysis. Three biological replicates were established per treatment at each time point, resulting in 18 samples in total. Each sample contained material from 15 individual plants. For RNA-seq analysis, hypocotyl tissue was cut and immediately flash frozen in liquid nitrogen. The preserved tissue was placed in long-term storage at −80 °C for RNA extraction.
The RNA prep Pure Plant Plus Kit (DP441, Tiangen, Beijing, China) was used to isolate mRNA directly from MM and BY hypocotyl tissue. Paired-end RNA-seq was carried out by Tiangen (Beijing, China) on the Illumina HiSeq 2000 platform. DEGs were identified by the following criteria: |log2| ≥ 1 and p value < 0.05. The DEGs between MM and BY were annotated on the basis of the B. rapa reference genome. The raw sequencing data have been deposited in the China National Center for Bioinformation under Accession No. PRJCA028426.
Quantitative real-time PCR
-
Total RNA was isolated with RNAiso Plus (Takara, Beijing, China). cDNA synthesis was performed with SuperScript Reverse Transcriptase (Invitrogen, Shanghai, China). A Roche thermal cycler (LightCycler 480, Roche, Switzerland) was used to perform qRT-PCR. BrACTIN was used as an internal control. Three independent experiments were performed with biological replicates containing material from approximately 15 seedlings per experiment.
Effector–reporter-based transactivation assay
-
pGreenII0800-LUC and pCambia2300 vectors were used for the effector–reporter-based transactivation assay as described previously[29,30]. The 2.0 kb promoter regions of BrA09EXPA5 in MM and BY were cloned and inserted into the pGreen II 0800-LUC vector (reporter). The coding sequences of BrA08BES1MM and BrA08BES1BY were subsequently cloned and inserted into the pCambia2300 vector (effector). Equal volumes of the suspensions were mixed and infiltrated into N. benthamiana leaves. The N. benthamiana leaves were placed in the dark for 24 h, and then transferred to the light for 48 h, and LUC activity was detected using a NightSHADE LB982 system (Berthold Technologies, Germany).
Generation of BrBES1 overexpression vectors and transgenic Arabidopsis plants
-
BrA08BES1MM and BrA06BES1MM cDNAs were cloned into the binary vector pCAMBIA2300. All the clones were confirmed by DNA sequencing. The resulting constructs 35S::BrA08BES1MM, 35S::BrA06BES1MM, and empty vector were transformed into Agrobacterium tumefaciens, strain GV3101. Transgenic plants were generated by the floral dip method and selected on ½ MS medium plus 80 mg/L kanamycin.
-
The turnip inbred line MM, with a long hypocotyl, and the Chinese cabbage inbred line BY, with a short hypocotyl, were chosen for studying hypocotyl length variation in B. rapa (Fig. 1a; Supplementary Fig. S1). Under normal light conditions, the hypocotyl of MM is longer than that of BY. The dynamic process of hypocotyl elongation were traced under 16 h light/8 h dark (LD) conditions to identify the key stage of hypocotyl length differentiation. The dynamic process covered a period of 18 d after sowing (DAS). The hypocotyl growth of MM was faster and lasted longer than that of BY (Fig. 1b, c). The hypocotyl length of MM at 5 DAS was already 2.14 cm longer than that of BY, indicating that the difference in hypocotyl length between MM and BY appeared very early during hypocotyl elongation (Fig. 1b). To investigate whether the difference in hypocotyl length between MM and BY resulted from an altered cell structure, the hypocotyl cells of MM and BY were observed at 3, 8, and 15 DAS (Fig. 1a, d). Compared with those of BY, the cells of MM exhibited differences in arrangement and vertical length (Fig. 1d). MM had an irregular surface containing cells that are long or short in the vertical dimension; however, BY had an orderly cell arrangement (Fig. 1d). In general, the length of MM cells was significantly greater than that of BY cells (Fig. 1e−g). These data demonstrated that the interspecific variation in hypocotyl length between MM and BY was linked to their difference in the vertical expansion of cells.
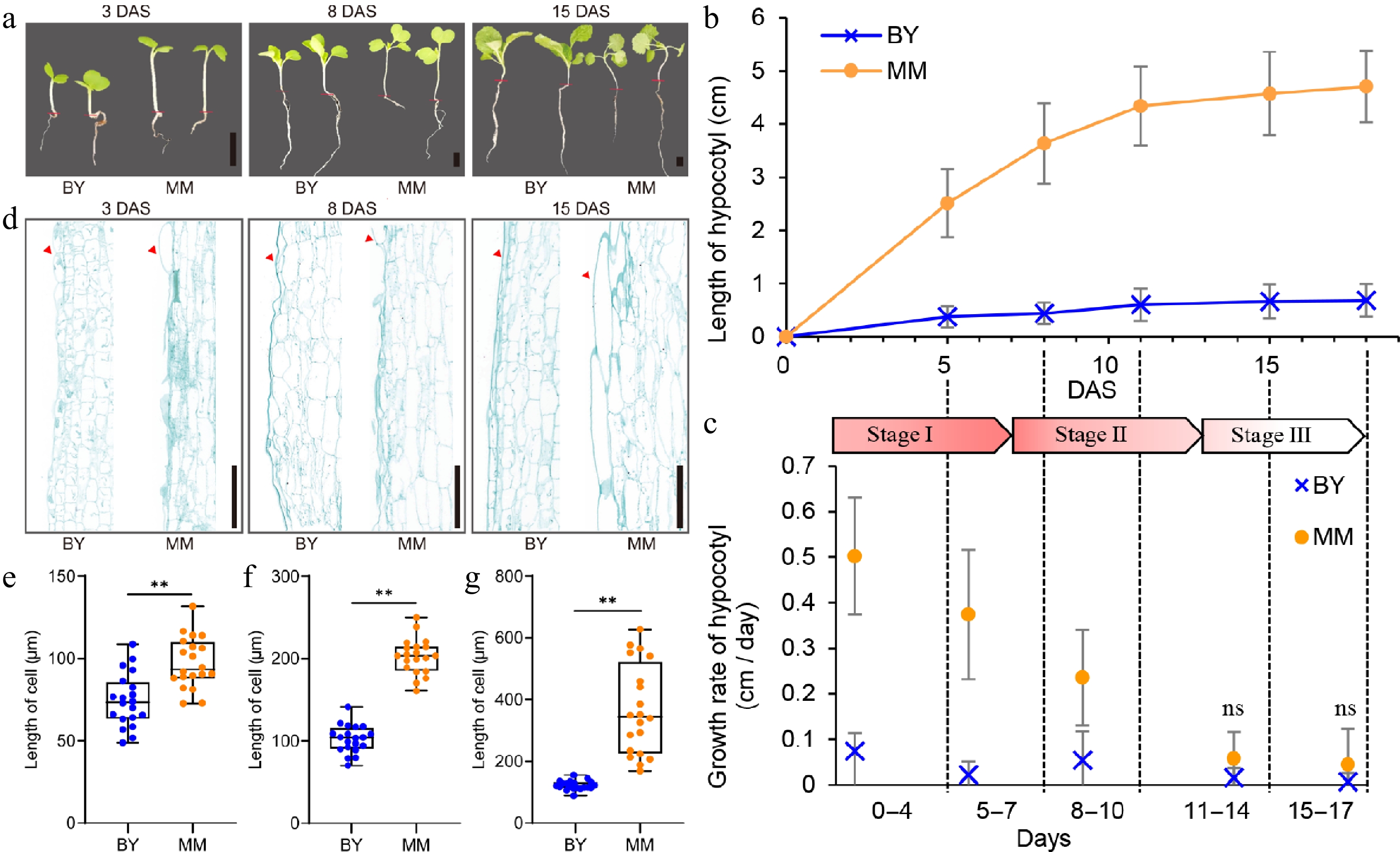
Figure 1.
Phenotypic differences between turnip (MM) and Chinese cabbage (BY). (a) Images of MM and BY seedlings at 3, 8, and 15 DAS. Scale bars = 1 cm. The red lines indicate the boundary between root and hypocotyl. (b) Dynamic comparison of hypocotyl length between MM and BY. Seeds were germinated overnight at 25 °C and then planted in the greenhouse under LD (16 h light/8 h dark) conditions. The error bars indicate the SDs; at least 15 plants of each inbred line were used. (c) Stage-specific growth rate of the hypocotyl. Stages I, II, and III represent the fast elongation stage (0–7 d), elongation stage (8–13 d), and ending stage (14–18 d), respectively. The arrow indicates the deadline on the timeline of each stage. (d) Longitudinal sections of hypocotyl cells from MM and BY. A partial image of cells from the middle section of the hypocotyl is shown. The red arrows indicate the epidermis. Scale bars = 500 µm. (e)–(g) Length of hypocotyl cells at (c) 3, (d) 8, and (e) 15 DAS. Significance level: ** p < 0.01.
Potential cues for hypocotyl length differentiation between MM and BY were identified through transcriptome analysis
-
To further study the variation in hypocotyl length in B. rapa via transcriptome analysis, the whole process was divided into three stages, namely, the fast elongation stage (0~7 DAS), elongation stage (8~13 DAS), and the ending stage (14~18 DAS), according to the growth rate of the hypocotyl (Fig. 1c). To determine the potential reasons for the difference in hypocotyl length between turnip and Chinese cabbage, temporal transcriptomic analysis was performed to investigate gene expression levels at a single time point in each stage during hypocotyl elongation. Time points of 3, 8, and 15 DAS were selected to represent the three stages of the early hypocotyl development on the basis of phenotype for a new batch experiment (Figs 1c, 2a). It was discovered that approximately 64% of the genes were expressed (the average over the three time points) in the hypocotyl of turnip, with approximately 10% of the genes upregulated or downregulated in MM compared with BY (Supplementary Fig. S2). The Venn diagram shows the differentially expressed genes (DEGs) between MM and BY at the corresponding time points (Fig. 2b). On the basis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis at all three time points, the DEGs upregulated in MM compared with BY were annotated mainly to the circadian rhythm-plant and plant hormone signal transduction pathways, both of which have been reported to be involved in hypocotyl elongation (Supplementary Fig. S3). The upregulated DEGs were as follows: PSEUDO RESPONSE REGULATOR (PRR7), PRR5, GIGANTEA (GI), and PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) in the circadian clock network; Gretchen Hagen3 (GH3), SMALL AUXIN UP RNAs (SAURs) in the auxin pathway; type-B ARABIDOPSIS RESPONSE REGULATORS (B-ARR) and A-ARR in the cytokine pathway; PYRABACTIN RESISTANCE1 (PYR) and SNF1-related kinase 2 (SnRK2) in the abscisic acid pathway; ethylene receptor (ETR); jasmonate ZIM-domain (JAZ) in the jasmonic acid pathway; and pathogenesis-related (PR-1) and Nonexpressor of PR 1 (NPR1) in the salicylic acid pathway (Fig. 2c). Moreover, the genes PHYTOCHROME B (PHYB) and AUX/IAA were upregulated in MM compared with BY only at 3 DAS. The trends in the expression of the BZR1/2 genes were consistent with the trend in hypocotyl elongation (Fig. 2c). To identify genes that might be related to the difference in hypocotyl length between MM and BY, the 2,041 genes that were differentially expressed at both 3 and 8 DAS timepoints but were not differentially expressed at 15 DAS (Fig. 2b) were studied, i.e., the DEGs during the hypocotyl fast elongation stage and elongation stage. The upregulated and downregulated genes among these 2,041 DEGs were annotated via Gene Ontology (GO) analysis (Supplementary Fig. S4). Notably, many of the downregulated DEGs were annotated to cell wall and hormone, which are highly related to BR; the upregulated DEGs were annotated to photosynthesis and photomorphogenesis terms (Supplementary Fig. S5). For example, BrrA07g232300, encoding a homolog of PACLOBUTRAZOL RESISTANCE 5 (PRE5), was identified among the photomorphogenesis-associated genes (Supplementary Table S2). PRE5 is a target gene of BZR1/BES1 that is required for promoting cell elongation[31]. This observation implies that the annotated biological processes might be associated with the difference in hypocotyl length between MM and BY.
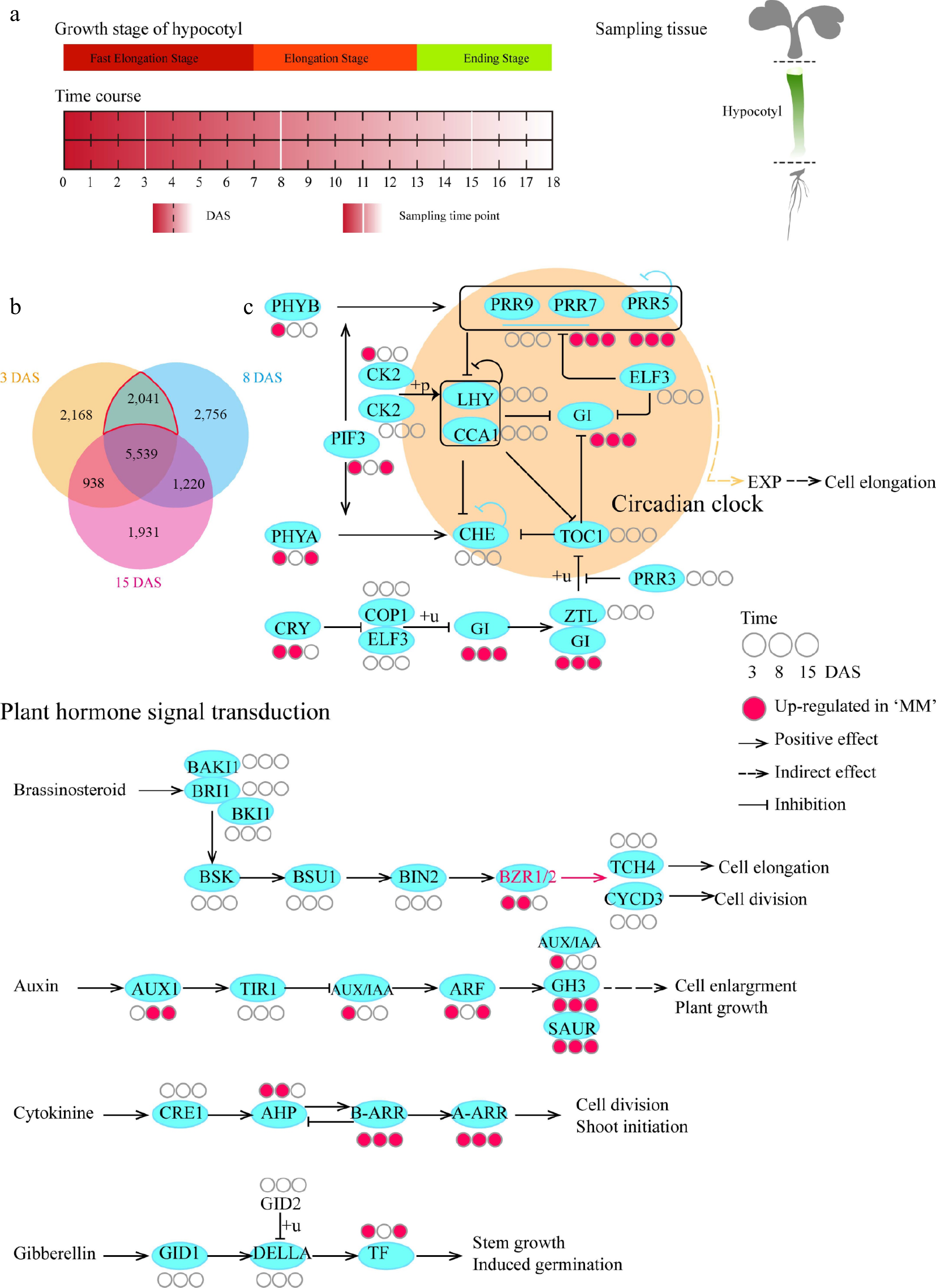
Figure 2.
Comparative temporal transcriptome analysis of hypocotyls between MM and BY. (a) Design of the transcriptome analysis. Seeds (MM and BY) were germinated overnight at 25 °C and planted in the greenhouse under LD conditions. Sampling was conducted for each line at 3, 8, and 15 DAS, covering the abovementioned three stages of hypocotyl growth. Tissue was sampled from the whole hypocotyl. For each line, three biological replicates at each time point were used. Each replicate contained material from at least 15 plants. (b) Venn diagram of DEGs. DEGs between MM and BY at 3, 8, and 15 DAS are shown in specific colors. The red line encircles the DEGs during the hypocotyl elongation stages. The upregulated and downregulated genes are not displayed separately. (c) KEGG enrichment analysis of the upregulated genes in the circadian rhythm and BR signaling pathways in turnip. The original maps showing the results of KEGG enrichment analysis are shown in Supplementary Table S1.
Comparison of BR sensitivity between MM and BY
-
As BRs promote hypocotyl elongation in B. rapa (pak choi)[24], and our transcriptome analysis revealed that BR-related pathways might be associated with the difference in hypocotyl length between MM and BY, we added brassinazole (Brz), an inhibitor of BR biosynthesis, to ½ MS medium to observe the germination and hypocotyl elongation of MM and BY. After 10 d, the hypocotyls of MM and BY plants growing on ½ MS medium supplemented with Brz were significantly shorter than those of the corresponding control plants (Fig. 3a, b). Control plants received the same volume of Brz solvent (DMSO) without the active compound. In addition to changing the hypocotyl length, Brz also led to a decrease in the cell length (Fig. 3c, d). The hypocotyl length of MM was decreased to different degrees when graded concentrations of Brz were added. The addition of a low concentration of Brz, 0.1 µM, repressed hypocotyl elongation in MM and BY to a similar degree, the addition of 1 µM Brz repressed hypocotyl elongation more strongly in MM than in BY, and MM and BY plants treated with 10 µM Brz presented no obvious hypocotyl elongation and exhibited a dwarf phenotype (Fig. 3e). These results indicate that BR promotes hypocotyl elongation through cell expansion in B. rapa, and MM is more sensitive to Brz than BY.
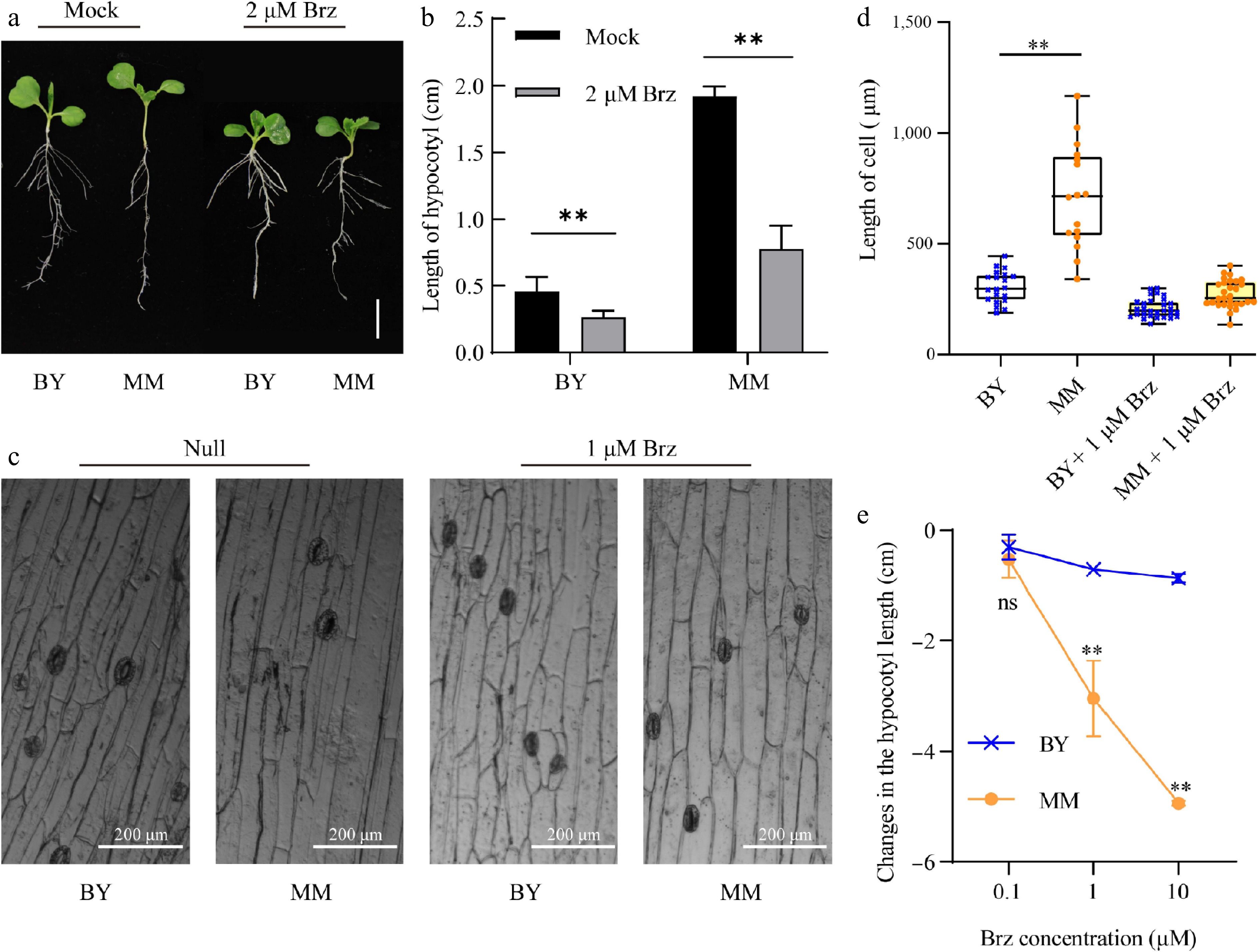
Figure 3.
Brz represses hypocotyl growth and hypocotyl cell elongation in MM and BY. (a) Morphological changes in MM and BY in response to Brz. Seeds were germinated on ½ MS culture medium supplemented with 2 µM Brz or control (mock) for 10 d under LD conditions. Bar = 1.9 cm. (b) Changes in the hypocotyl length of MM and BY in response to Brz. Seeds were germinated on ½ MS culture medium supplemented with 2 µM Brz or control medium (mock) for 10 d under LD conditions. The error bars indicate the SDs; n = 6. Significance level: ** p < 0.01. (c) Visualization of epidermal cells in the hypocotyls of MM and BY treated with Brz. (d) Changes in the epidermal cell length in the hypocotyls of MM and BY in response to Brz. Significance level: ** p < 0.01. (e) Changes in the hypocotyl lengths of MM and BY under treatment with gradient concentrations (0.1, 1, and 10 µM). The error bars indicate the SDs; n = 6.
Comparison of BR biosynthesis between MM and BY
-
Given the differential sensitivity of MM and BY to Brz treatment, it was hypothesized that BR biosynthesis pathways may also differ between the two accessions. We subsequently quantified endogenous brassinosteroid levels at two critical developmental stages of hypocotyl elongation, the early (6 DAS) stage and late (15 DAS). Five BR compounds belonging to three BR biosynthesis pathways were identified (Fig. 4a, b): 6-deoxocastasterone (6-deoxoCS), typhasterol (TY), 28-norcastasterone (28-norCS), castasterone (CS), and brassinolide (BL). At 6 DAS, the abundances of BL and the intermediate TY in MM were slightly greater than those in BY. In contrast, the abundances of the intermediate 6-deoxoCS and CS in MM were lower than those in BY, suggesting that the BR synthesis pathways in MM and BY are different. At 15 DAS, the abundances of BL in BY and MM hypocotyls were decreased, and the abundances of the intermediates were also low in BY. However, the abundances of the intermediates TY, CS, and 28norCS remained high in MM (Fig. 4b). Approximately 14 genes involved in BR biosynthesis were identified in the B. rapa genome via ortholog analysis on the basis of the Arabidopsis BR biosynthesis pathways (Supplementary Table S3), and the expression levels of these genes were compared between MM and BY. The expression levels of some homologs in turnip and Chinese cabbage were inversely related (Supplementary Fig. S6). As an example, AtDWF4 encodes a cytochrome P450 monooxygenase in the BR biosynthesis pathway and can be repressed by Brz[32,33]. In B. rapa, two homologs of AtDWF4, namely, BrA01DWF4 and BrA09DWF4, were identified, and their relative expression levels were determined via quantitative real-time PCR (qRT‒PCR) (Fig. 4c). The expression level of BrA01DWF4 in BY was significantly greater than that in MM, whereas the expression level of BrA09DWF4 in MM was greater than that in BY (Fig. 4c). The results revealed that BRs were continuously synthesized in hypocotyl tissue during the hypocotyl elongation stage of B. rapa. In addition, BRs may be involved in other processes when hypocotyl elongation in MM is nearly complete.
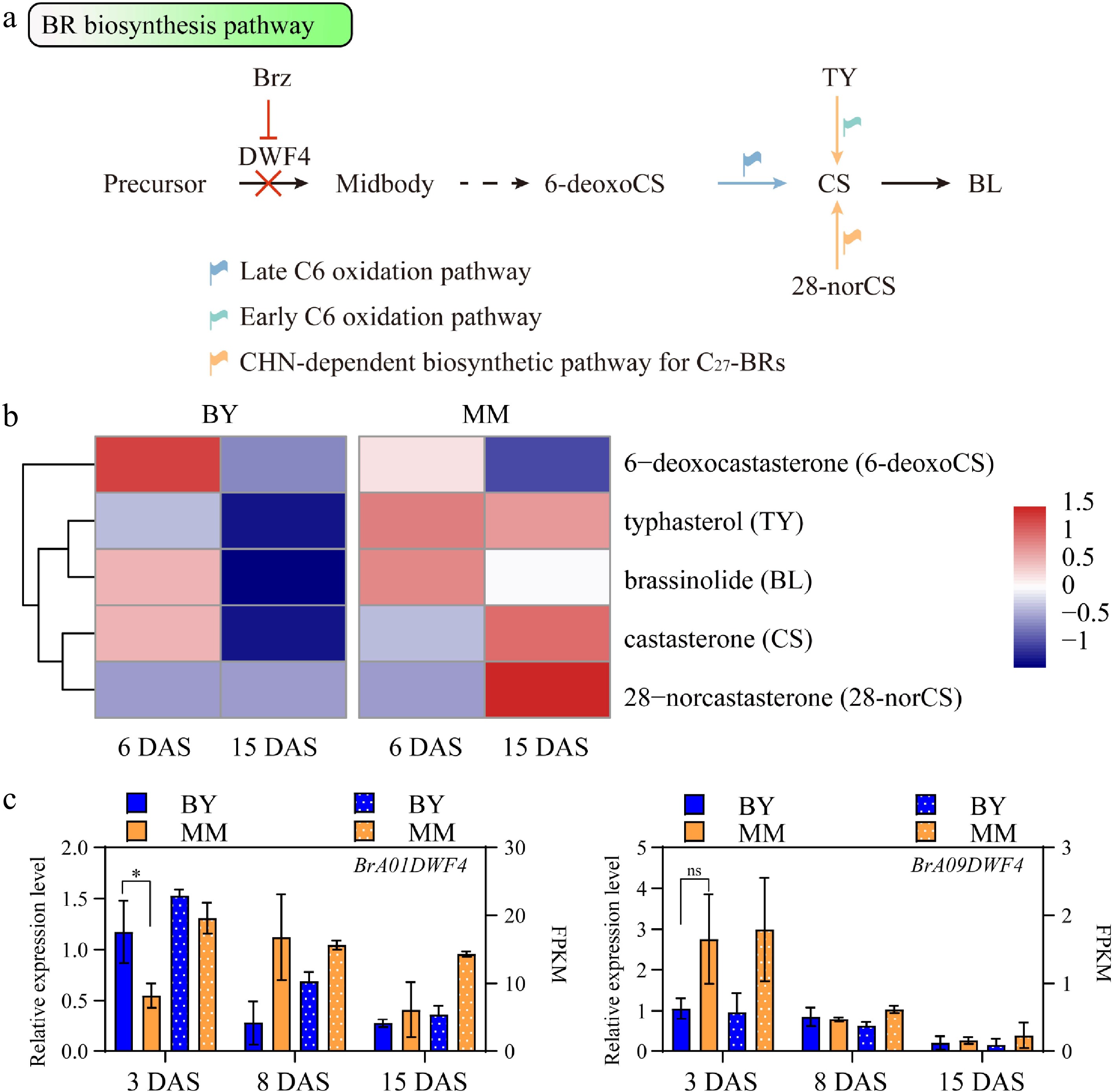
Figure 4.
Comparison of the BR biosynthesis pathway between MM and BY. (a) BR biosynthesis pathway in plants. The likely preferential pathways in MM and BY are indicated by orange and blue arrows, respectively. The dashed arrow indicates more than one step. (b) Heatmap of the BR concentrations in the hypocotyls of MM and BY at 6 and 15 DAS. (c) Expression levels of BrA01DWF4 and BrA09DWF4 determined via qRT‒PCR and RNA-seq in the hypocotyl of B. rapa at 3, 8, and 15 DAS. Significance level: * p < 0.05 by an unpaired t test; ns p value > 0.05 by an unpaired t test.
Differences in the BR signaling pathway between MM and BY
-
Thus far, experimental evidence indicates that the differences in BR sensitivity may lead to the interspecies variation in hypocotyl length in B. rapa. We further hypothesized that BR signal transduction is involved in the variation in hypocotyl length. Therefore, we grew MM and BY on ½ MS medium supplemented with 24-epibrassinolide (24-epiBL) to compare the BR signal transduction ability. Hypocotyl elongation in MM was greater than that in BY under 24-epiBL treatment (Fig. 5a). The hypocotyl was significantly longer in MM plants treated with 100 nM 24-epiBL than in the control MM plants, whereas there was no significant difference in hypocotyl length between treated and control BY plants (Fig. 5b). The maximum hypocotyl elongation in both MM or BY was the same as that without Brz treatment, indicating that the level of BR signal transduction activity in MM and BY was accurately reflected (Fig. 5b). These data prove that MM is more sensitive to BR than BY.
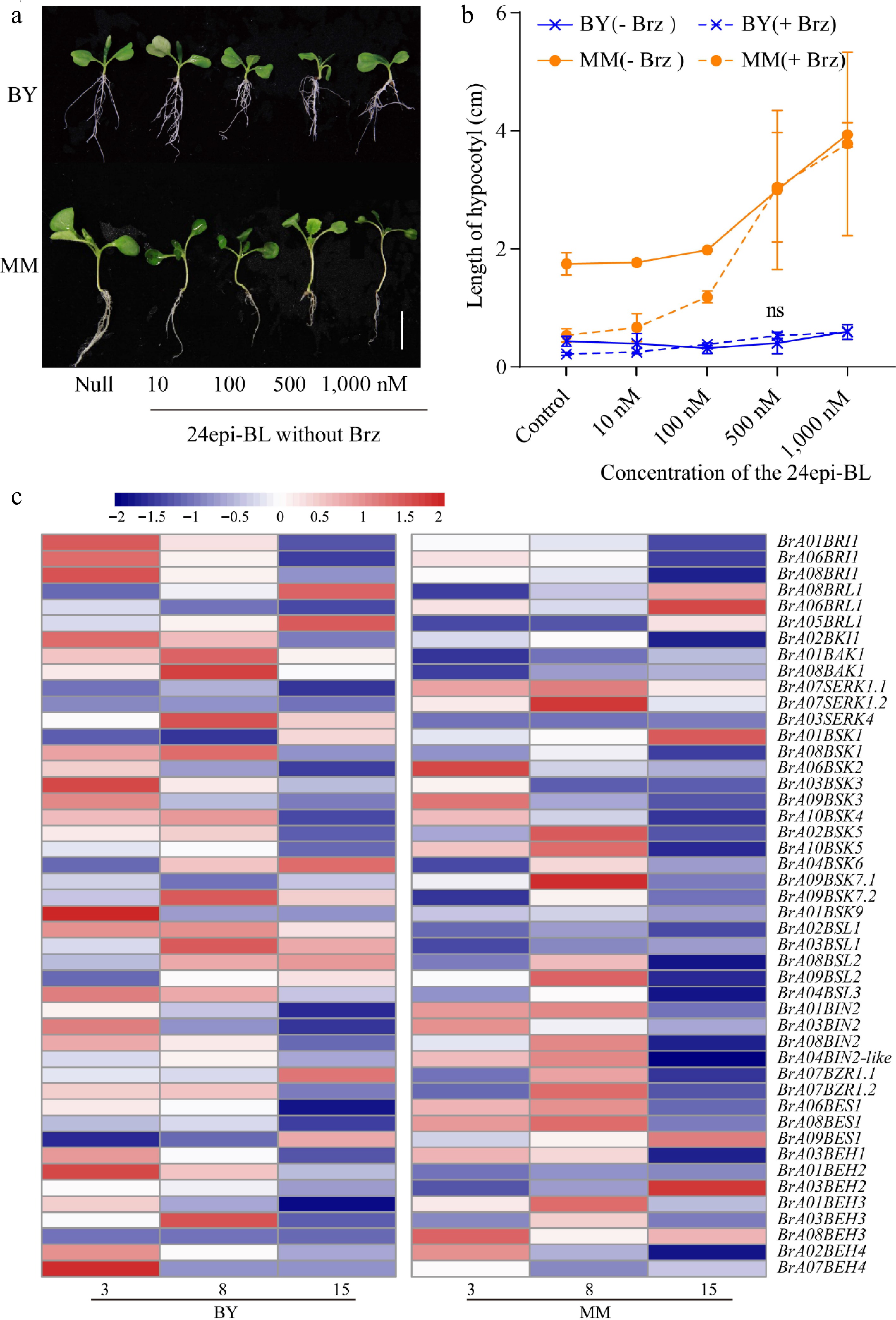
Figure 5.
The response of MM to BR signaling is stronger than that of BY. (a) Morphological changes in MM and BY in response to 24epi-BL. Seeds were germinated on ½ MS culture medium supplemented with gradient concentrations of 24epi-BL (10, 100, 500, and 1,000 nM) and untreated medium (blank application) under LD conditions for 10 d. Bar = 1.9 cm. (b) Measurement of hypocotyl elongation in MM and BY seedlings under treatment with BL with or without Brz. Seeds were germinated on ½ MS culture medium (containing 0 or 1 µM Brz) supplemented with gradient concentrations of 24Epi-BL (10, 100, 500, and 1,000 nM) under LD conditions, and the hypocotyl length was measured at 10 DAS. At least five seedlings were used per replicate. The error bars indicate the SDs; n = 3. (c) Heatmap of the transcript levels of BR signaling pathway genes in the hypocotyls of MM and BY at 3, 8, and 15 DAS.
BR signaling pathway genes regulate hypocotyl length by directly activating the transcription of cell expansion genes
-
Next, the BR signaling pathway genes were identified via MapMan. A total of 47 genes homologous to Arabidopsis BR signaling genes were identified in B. rapa (Supplementary Table S4). These genes were duplicated or lost in B. rapa relative to Arabidopsis. The expression levels and amino acid sequences of these genes were subsequently compared between turnip and Chinese cabbage. Sequence alignment analysis revealed that the genomic sequences of most of the genes differed between turnip and Chinese cabbage, possibly leading to functional divarication in these genes (Supplementary Table S4, Supplementary Fig. S7). Interestingly, many genes positively regulating hypocotyl length, except for BZR1/2, presented higher expression levels in BY than in MM (Figs 2c, 5c). Previous studies have shown that BZR family members redundantly regulate vegetative tissue growth in Arabidopsis[34]. In B. rapa, two, three, and eight homologs of BZR1, BES1, and BEH, respectively, in the BZR family were identified. Transcriptome analysis revealed that among BZRs, the BrA08BES1 gene had the highest expression level in B. rapa. To validate the transcriptome data, qRT-PCR analysis of BrA08BES1 in MM and BY hypocotyls was performed at 3, 8, and 15 DAS. The results confirmed that BrA08BES1 was significantly more highly expressed in MM than in BY at 3 and 8 DAS (Supplementary Fig. S8), thereby verifying the RNA-seq data and correlating with the hypocotyl elongation phenotype.
The promoter and coding sequences (CDSs) of BrA08BES1 were sequenced and compared between MM and BY, and the most obvious variation identified was an 80 bp insertion at −47 bp of BrA08BES1BY relative to BrA08BES1MM. Moreover, the positions of the start codons of BrA08BES1BY and BrA08BES1MM were different. The start codon of BrA08BES1BY is located at +235 bp relative to that of BrA08BES1MM. If the transcription of BrA08BES1BY is initiated at the same location as BrA08BES1MM transcription, translation will be terminated at +151 bp because a 1 bp deletion mutation in BY will cause a frameshift. Therefore, the promoter region of BrA08BES1BY is 324 bp longer than that of BrA08BES1MM (Supplementary Fig. S9). BES1 was reported to promote hypocotyl elongation through direct upregulation of a K+ channel gene (KAT), SMALL AUXIN UP RNAs (SAURs), and EXPA in Arabidopsis[11]. To prove the function of BZRs in activating the transcription of cell expansion genes in B. rapa, the expansin gene BrA09EXPA5, which has complete sequence conservation between MM and BY and is expressed at higher levels in MM than in BY, was chosen (Fig. 6a). The ability of BrA08BES1 to activate BrA09EXPA5 transcription was confirmed by a luciferase (LUC) effector–reporter assay in Nicotiana benthamiana epidermal cells (Fig. 6b). Although the N-terminal amino acid sequence of BrA08BES1MM is longer than that of BrA08BES1BY (Supplementary Fig. S10), the LUC intensity generated via BrA08BES1MM- and BrA08BES1BY-mediated activation of pBrA09EXPA5-LUC was similar, indicating that the functions of these genes in regulating hypocotyl length are generally the same (Fig. 6c). To provide direct genetic evidence for the role of BrBZRs in hypocotyl elongation, transgenic Arabidopsis plants overexpressing BrA08BES1MM and BrA06BES1MM under the control of the 35S promoter were generated. The hypocotyls of 35S::BrA08BES1MM transgenic plants were slightly but significantly longer than those of the empty vector controls (Supplementary Fig. S11a, S11c). Meanwhile, 35S::BrA06BES1MM plants also exhibited a subtle increase in hypocotyl length, although this trend was not statistically significant (Supplementary Fig. S11b, S11c). These preliminary results indicate that BrA08BES1MM plays a partial, promotive role in regulating hypocotyl elongation.
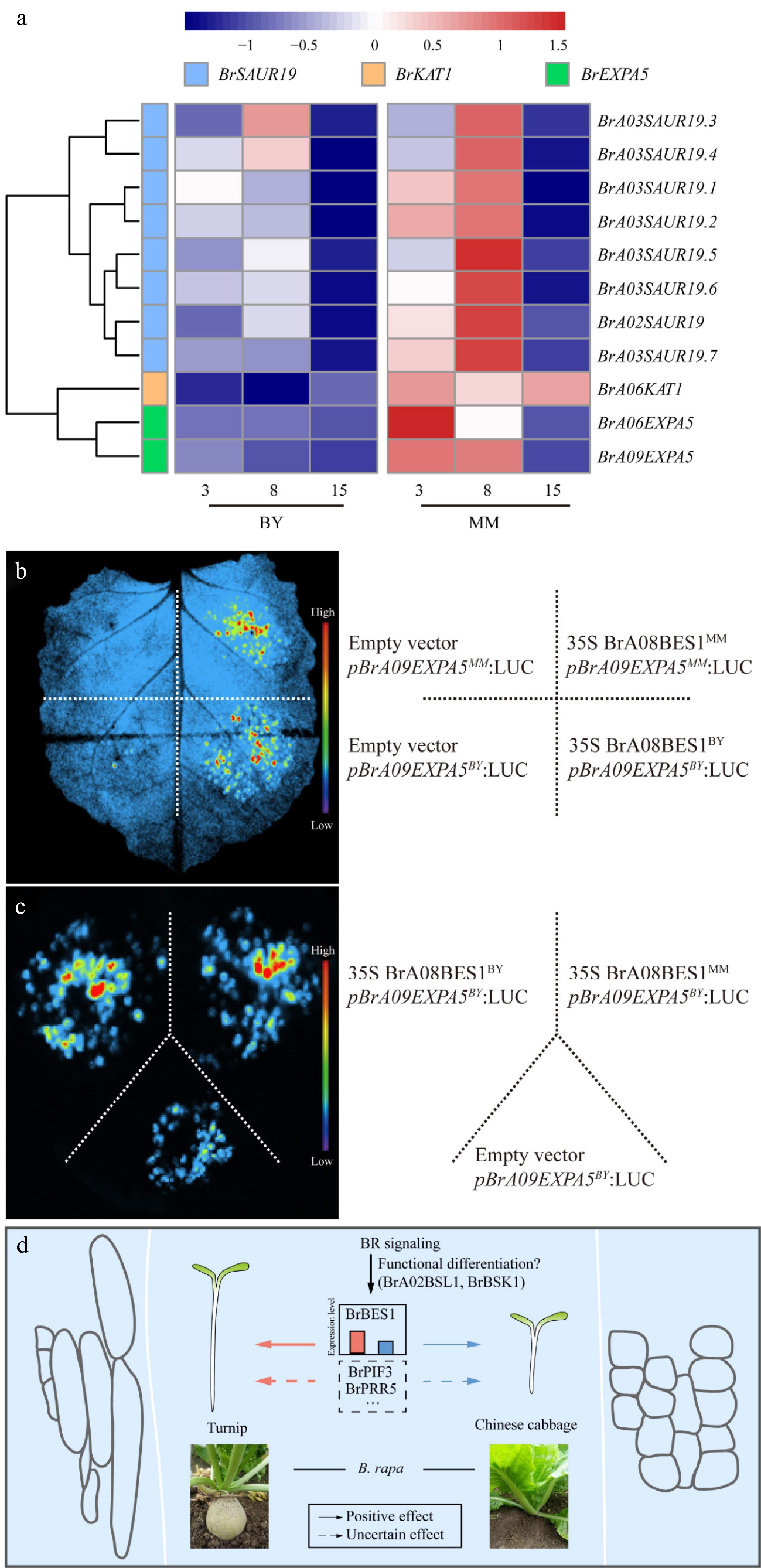
Figure 6.
BR signaling transcription factors activate the transcription of cell expansion genes, which are specifically expressed during hypocotyl elongation in MM and BY. (a) Heatmap of the transcript levels of the cell expansion genes in the hypocotyls of MM and BY at 3, 8, and 15 DAS. (b) BrA08BES1 activates the transcription of the gene BrA09EXPA5 in N. benthamiana epidermal cells, as shown by an effector–reporter-based transactivation assay. The empty vector and pBrA09EXPA5 were injected as negative controls. (c) BrA08BES1BY and BrA08BES1MM activate the transcription of the gene BrA09EXPA5BY. The empty vector and pBrA09EXPA5BY were injected as negative controls. (d) BR signaling pathway differences contribute to the variation in hypocotyl elongation between turnip and Chinese cabbage.
It is therefore hypothesized that the natural variation in the promoter region of BrA08BES1 between the two B. rapa subspecies contributes to its differential expression, which in part underlies the observed divergence in hypocotyl length, rather than through functional divergence of the protein itself. Collectively, the present findings support the model that the natural variation in hypocotyl elongation is influenced, at least in part, by differential expression of BZR transcription factors within the BR signaling pathway.
-
Brassica species display extensive morphological diversity[20−22]. Turnip is characterized by a swollen taproot, while Chinese cabbage forms a large leafy head. A particularly striking phenotypic difference between turnip and Chinese cabbage is their hypocotyl length variation during early growth and development. Hypocotyl elongation is an essential process in plant morphogenesis and is regulated by the coordinated action of environmental stimuli and endogenous factors[1,6,35,36]. Turnip and Chinese cabbage are closely related but show big interspecies differences in terms of hypocotyl length; thus, they are very suitable for exploring the mechanism underlying interspecies differences in hypocotyl regulation. A major aim of this work was to reveal the genetic and physiological differences associated with the difference in hypocotyl length between turnip and Chinese cabbage. This issue was addressed via transcriptome analysis of two inbred lines, MM (turnip) and BY (Chinese cabbage), during the hypocotyl elongation process. Photomorphogenesis and skotomorphogenesis are known to involve hypocotyl elongation, and many DEGs between MM and BY have been verified. In addition, circadian rhythm, photosynthesis, hormone biosynthesis, signal transduction and metabolism, and cell wall modification might contribute to the difference in hypocotyl length between MM and BY. Another comparative study of hypocotyls between turnip and Chinese cabbage revealed no obvious differences in either morphological characteristics or vascular development (as observed in cross-sections) during 0–12 DAS, based on cytological observations[37]. The results of this study suggest that the hypocotyls have not yet started lateral expansion, supporting our conclusion that the hypocotyls remain in the elongation phase from 0–14 DAS.
Differences in BR biosynthesis between turnip and Chinese cabbage
-
In the present study, differences were observed in both the expression levels of genes involved in the BR biosynthesis pathway and the endogenous BR levels between turnip and Chinese cabbage. Although the genes in the BR biosynthesis pathway exhibited differential expression levels between MM and BY, the presence of multiple gene copies in the B. rapa genome complicates the establishment of clear relationships between gene expression and BR content. Heatmap shows only five genes—CONSTITUTIVE PHOTOMORPHOGENIC DWARF (BrA02CPD), BrA01DWF4, BrA09DWF4, ROTUNDIFOLIA3 (BrA03ROT3), and BRASSINOSTEROID-6-oxidase 2 (BrA06BR6ox2)—that were positively associated with hypocotyl elongation (Supplementary Fig. S6). Additionally, five BRs in the hypocotyl of B. rapa: 6-deoxoCS, TY, 28-norCS, CS, and BL were detected and quantified A particularly notable difference was observed in the contents of TY and CS. TY was substantially more abundant in MM than in BY, whereas the opposite was true for CS. Both CS and BL, which are C28-BRs, are known to be highly biologically active[38]. The higher BL content in MM compared to BY suggests that the overall levels of biologically active BRs might be comparable between the two varieties. C28-BRs are synthesized from campesterol (CR) via a campestanol (CN)-dependent pathway and a CN-independent pathway[39,40]. Furthermore, the notably higher levels of TY, CS, and 28-norCS in MM at DAS15 may contribute to lateral expansion. 28-norCS, a C27 counterpart of CS, is synthesized from cholesterol via a mechanism similar to that mediating the synthesis of CS from CR[40]. On the basis of these findings, we speculate that in MM, BR biosynthesis is preferentially conducted via the early C6 oxidation pathway and CHN-dependent biosynthetic pathway to yield C27-BRs; in contrast, the late oxidation pathway is preferentially activated in BY (Fig. 4b).
Differences in BR perception between turnip and Chinese cabbage
-
Here, it was found that turnip (MM) is more sensitive to BR than Chinese cabbage (BY), suggesting differences in the BR signaling pathway between turnip and Chinese cabbage. The differences originate from variations in gene function, expression levels, and post-transcriptional control.
Core components of the BR signaling pathway were identified and their sequence variations compared between turnip and Chinese cabbage. Substantial sequence divergence exists for several genes between these species. Additionally, our analysis found that Brassica crops have lost the gene BRI1 suppressor 1 (BSU1, AT1G03445) compared to Arabidopsis. Thus, functional divergence of signaling components is an important factor underlying the differential BR response.
At the transcriptional level, most BR signaling pathway genes exhibit upregulation in BY. This pattern, however, contrasts with the greater BR sensitivity of MM, presenting a paradox. Strikingly, it was noticed that the expression levels of seven out of nine BZR family genes were highly expressed in MM. These include BrA06BES1, BrA08BES1, BrA09BES1, and BrA08BEH3, which were up-regulated in MM both at 3 and 8 DAS; and BrA07BZR1.1, BrA07BZR1.2, and BrA01BEH3, which were up-regulated specifically at 8 DAS. The temporal expression patterns of BrA06BES1 and BrA08BES1 were related to the dynamics of hypocotyl elongation. In the present study, BrA08BES1MM and BrA08BES1BY were shown to activate conserved cell expansion genes.
Furuya et al. provided strong evidence indicating that BES/BZR homologs have a competitive relationship in the regulation of downstream genes[15]. In tomato, five BES1 family members exhibit transcriptional activation activity, whereas the remaining two function as transcriptional repressors; SlBES1.8 is expressed specifically in flowers, whereas the other BES1 family members are expressed ubiquitously in all organs[41]. The preliminary results demonstrate a mild yet significant promotive effect of BrA08BES1MM on hypocotyl elongation, mirroring the phenotype of 35S::AtBES1[42]. Meanwhile, other BZR1/BES1 homologs in the B. rapa genome might also contribute to the regulation of hypocotyl elongation, potentially redundantly or synergistically.
BES1/BZR1 family transcription factors regulate plant development via BR-dependent and BR-independent pathways[43]. Their activity is modulated by multiple mechanisms. In addition to BIN2-mediated phosphorylation of BES1, which inhibits its DNA-binding activity toward BR-responsive target promoters and its transcriptional activity through impaired multimerization[8]. Photoreceptors interact with BSE1 to regulate hypocotyl elongation. The cryptochromes (CRY1)-BES1 interaction leads to both inhibition of the DNA binding activity of BES1 and the repression of its target genes[44]. Recent research has shown that green light promotes the DNA binding activity of BES1, a master transcription factor in the BR pathway, thus regulating gene transcription to promote hypocotyl elongation[45]. TCP8 interacts directly with the master BR regulators BZR1 and BZR2 in multiple expression systems[46]. Therefore, identifying the upstream regulators of BrA08BES1 and the key functional sites underlying its role in hypocotyl variation will greatly advance our understanding of this regulatory network.
Application of genes related to the regulation of hypocotyl elongation to mechanized harvesting in agricultural production
-
In crop breeding, increased crop yield and improved production efficiency are consistently the goals of breeders, and modulating genes related to plant growth is a rapid way to achieve these goals. Hypocotyl length affects the relative height of aboveground tissue with respect to the ground and is an important agronomic characteristic influencing mechanized production efficiency. During the mechanized harvesting of leafy vegetables, maintaining an appropriate distance between the aboveground nutrient organs and the ground is beneficial for reducing damage inflicted by the harvesting arm to the nutrient organs of the vegetable, reducing yield loss, and increasing storage time[23]. In addition, hypocotyl length is an important component of plant height and affects the lodging and yield of leafy vegetables. Therefore, understanding the molecular mechanism of hypocotyl elongation in crop plants is beneficial for breeding cultivars with strong seedlings or suitable for mechanized harvesting.
The present study revealed a genetic basis for BR signaling in turnip and Chinese cabbage and showed the differences in the control of the physiological processes of hypocotyl elongation (Fig. 6d). Treatment with different concentrations of BrBZRs induced different activation profiles of downstream genes to control cell expansion, contributing to hypocotyl length variation in B. rapa. Research on the genetic drivers underlying the substantial degree of morphological variation between B. rapa subspecies will prompt further evolutionary and functional studies. The present findings could be applied in agricultural production via modulation of the key genes to meet the needs for crop production.
This work was financially supported by grants from the National Natural Science Foundation of China (Grant No. 32272714), the Innovation and Capacity-Building Project of BAAFS (KJCX20251402, KJCX20230126, KJCX20230403), the Collaborative Innovation Program of the Beijing Vegetable Research Center (XTCX202402), the Innovation and Development Program of Beijing Vegetable Research Center (KYCX202504), the Post Doctor Foundation of Beijing (Grant No. 030220273), and the Young Talent Award of Beijing Agricultural and Forestry Science and Science Innovation Program (KYCX202001-07). We are grateful to members of the Chinese cabbage laboratory, particularly Xiaoyun Xin, Mei Zheng, and Xinyan Liu, for useful discussions and technical support.
-
The authors confirm contribution to the paper as follows: study conception and design: Wang Z, Li H; data collection: Li H, Li P, Wu H, Wang L, Wu Y; analysis and interpretation of results: Li H, Li P, Xin H; draft manuscript preparation: Wang Z, Li H, Li P. All authors reviewed the results and approved the final version of the manuscript.
-
The data that support the findings of this study are available in the China National Center for Bioinformation. These data were derived from the following resources available in the public domain: Accession No. PRJCA028426.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 The upregulated genes in the circadian rhythm and BR signaling pathways in turnip were subjected to KEGG enrichment analysis.
- Supplementary Table S2 Gene Ontology annotations of the overlapping DEGs at 3 DAS and 8 DAS but not at 15 DAS.
- Supplementary Table S3 BR biosynthesis pathway genes in B.rapa.
- Supplementary Table S4 BR signaling pathway gene identification via MapMan.
- Supplementary Fig. S1 Image of MM and BY growing under natural conditions at seedling (5 DAS) and premature stages.
- Supplementary Fig. S2 Statistics of gene expression at temporal transcriptome datasets.
- Supplementary Fig. S3 KEGG enrichment of DEGs between turnip MM and Chinese cabbage BY hypocotyl development at 3 DAS, 8 DAS and 15 DAS. Up: DEGs up-regulated in MM compared to BY.
- Supplementary Fig. S4 Number of genes both up-regulated and down-regulated at 3 DAS and 8 DAS in MM compared to BY.
- Supplementary Fig. S5 Go ortholog annotation of DEGs during hypocotyl elongation stages.
- Supplementary Fig. S6 Genes expression level of BR biosynthesis pathway in B. rapa.
- Supplementary Fig. S7 Alignment of BR signaling pathway proteins between MM and Chiifu.
- Supplementary Fig. S8 Relative expression levels of BrA08BES1 in the hypocotyls of MM and BY at 3 DAS, 8 DAS, and 15 DAS as determined by qRT-PCR. ** p < 0.01.
- Supplementary Fig. S9 Alignment of the promoter sequence and CDS of gene BrA08BES1 between MM and BY.
- Supplementary Fig. S10 Alignment of the BrA08BES1 protein sequences between MM and BY.
- Supplementary Fig. S11 BrA08BES1MM and BrA06BES1MM promote hypocotyl elongation in Arabidopsis.
- Copyright: © 2026 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li H, Li P, Wu H, Xin H, Wang L, et al. 2026. Differences in BR perception contribute to hypocotyl length variation in Brassica rapa. Vegetable Research 6: e006 doi: 10.48130/vegres-0025-0047
Differences in BR perception contribute to hypocotyl length variation in Brassica rapa
- Received: 30 May 2025
- Revised: 31 October 2025
- Accepted: 24 November 2025
- Published online: 04 February 2026
Abstract: Brassica crops exhibit variations in hypocotyl length under natural conditions, and the underlying genetic mechanism remains to be explored. Here, comparative transcriptome analysis revealed that numerous genes differentially expressed between turnip (MM) and Chinese cabbage (BY) at the hypocotyl elongation stage were associated with brassinosteroid (BR)-related pathways. Exogenous application of 24-epibrassinolide (24-epiBL) showed that turnip was more sensitive to 24-epiBL than Chinese cabbage. BrA08BES1, an ortholog of the BRI1-EMS-SUPPRESSOR 1 (AtBES1)/BRASSINAZOLE-RESISTANT 2 (AtBZR2) gene encodes a key transcription factor in the BR signaling pathway and exhibited the highest expression among BrBZR family members in turnip. Effector-reporter-based transactivation assays confirmed that BrA08BES1 activates the transcription of the potential cell expansion gene EXPANSINS 5 (BrA09EXPA5) in both MM and BY. The expression levels of BrA08BES1 and BrA09EXPA5 in turnip are higher than in Chinese cabbage, which is associated with their hypocotyl length. Furthermore, preliminary results from overexpressing BrA08BES1MM in Arabidopsis demonstrate that this gene plays a partial, promotive role in hypocotyl elongation. The present research reveals fundamental differences in BR signaling between turnip and Chinese cabbage. These findings will facilitate future evolutionary and functional studies of hypocotyl phenotype in B. rapa and improve mechanized production efficiency via molecular breeding.
-
Key words:
- Brassica rapa /
- Hypocotyl elongation /
- Cell expansion /
- Transcriptome /
- Brassinosteroid /
- Turnip /
- Chinese cabbage













