-
Goats are considered to be an economically important species, providing essential products for human consumption such as meat, milk, and fiber[1]. Coat color and cashmere traits are particularly significant characteristics, reflecting breeding progress and influencing the market value of goat products[2]. Cashmere, renowned for its softness and warmth, is a highly valued luxury fiber product that contributes significantly to the global economy[3]. Its production is a complex biological process occurring within the skin follicles and is regulated by a wide range of proteins. The quality, quantity, and color of cashmere are influenced by both genetic and environmental factors[4]. With the growing awareness of environmental issues and increasing consumer demand for natural products, the breeding of natural-colored goat breeds has become important[5]. Understanding the genetic basis of these traits is crucial for improving breeds and enhancing farmers' economic benefits.
Coat color is a key basis for breed identification and strain delineation in breeding research. In addition, it plays important roles in camouflage, predator avoidance, social communication, and courtship behavior[6,7]. The formation of different coat colors is determined by the number of melanocytes and the type of melanin in the skin[8]. Since the early 20th century, researchers have investigated the genetic mechanisms underlying coat color inheritance in goats. Several genes, including MITF, ASIP, TYR, TYRP1, and DCT, have been identified as major regulators of this trait. MITF is a key transcription factor involved in melanocytes' development, proliferation, and survival[9]. The ASIP gene is highly correlated with a light coat color. Agouti signaling protein (ASIP) competes with α-melanocyte-stimulating hormone (α-MSH) for binding to the melanocortin-1 receptor (MC1R), blocking the MC1R-induced cyclic adenosine monophosphate–protein kinase A (cAMP/PKA) signaling pathway, then reduces tyrosinase content and eumelanin synthesis, resulting in the dilution of coat color[10,11]. Jan Henkel reported that different ASIP alleles show diverse coat colors in Valais goats[11]. TYR, TYRP1, and DCT are critical for coat pigmentation and their different expression levels are linked to diversity in the phenotype. Mutations in the TYR gene's coding region can produce a mutant protein that is degraded by proteases, triggering a loss of function that impairs melanin synthesis and causes dilution of coat color. For instance, a c.138T>A mutation in Exon 1 of the TYR gene leads to a white coat in minks[12]. TYRP1 and DCT show elevated expression in the black skin regions of Boer × Macheng F1 hybrid goats[13]. In Copperneck goats, the introgression of a mutant TYRP1 genotype has been identified as a major contributor to brown pigmentation[11]. Consistent with this, Bat et al. also reported a significant association between DCT and the brown coat phenotype in goats[14]. The complex interplay among these genetic determinants continues to be a subject of active research that is fundamental to understanding the molecular basis of coat color variation.
The longer and finer fiber is the fundamental characteristic that distinguishes cashmere goats from other types of goats, serving as a key determinant of the fiber's quality and economic value[15]. Previous studies have reported that the quality and yield of fiber in goats are influenced by traits such as follicle development, follicle density, fiber diameter, and fiber length[16,17]. For instance, Wang et al. integrated whole-genome sequencing and skin transcriptomic data from Tibetan antelope and Siberian ibex, identifying Type IV collagen genes (e.g., COL4A2, COL4A4) and integrin genes (e.g., ITGA2, ITGA4) as potential candidates involved in the development of cashmere fiber[18]. Moreover, Zhao et al. conducted transcriptome sequencing on the skin of Dazu black goats and Inner Mongolian cashmere goats, identifying multiple mRNAs and noncoding RNAs associated with hair follicle development. They then constructed a competing endogenous RNA (ceRNA) regulatory network[19]. Zhang et al. investigated follicle density in goats and identified GJA1 and GPRC5D as candidate genes[20]. Similarly, Qin et al. performed transcriptome sequencing on skin samples from Liaoning cashmere goats with varying fiber diameters, identifying 16 genes associated with the fineness of cashmere[21]. Regarding fiber length, genes including FGF5, HOXC8, and KAP6-1 have been annotated[22−24]. Moreover, studies have indicated that correlations among cashmere-related traits, longer hair, and down fibers are generally associated with better fiber quality. This suggests that fiber length could serve as a selection criterion for enhancing other economic traits[17].
A deeper understanding of the genetic basis of production-related phenotypic traits can help improve production efficiency and breeding programs. Therefore, this study aimed to identify core genes associated with coat color and fiber length by conducting phenotypic and transcriptomic analyses of skin samples from four goat breeds, and attempted to investigate the genetic and molecular mechanisms that underlie these economically important traits.
-
Fiber and skin samples were collected from four goat breeds: Dazu black goat (DBG, n = 30), Yudong black goat (YBG, n = 30), Banjiao goat (BJG, n = 30), and Inner Mongolian cashmere goat (IMCG, n = 30) at the Southwest University Farm. The animals were fed with a standard artificial diet consisting of alfalfa, barley, and wheat straw. The feed's composition fully complies with the nutritional guidelines for goats established by the National Research Council (2007). All of the experimental goats were adult, healthy, and selected at random. Fiber and skin samples were collected in December, fiber samples (coarse hair and cashmere) were collected from a 10 cm × 10 cm patch on the side of the body located behind the shoulder of adult goats by shaving very closely to the skin with an electric razor. Skin samples were collected from the same area immediately after slaughter. The fiber samples were washed with absolute alcohol and then air-dried in a draught cupboard. The skin samples were divided into two parts: One was frozen in liquid nitrogen, and the other was preserved in paraformaldehyde.
Measurement of pigmentation traits and fiber length
-
Melanin content was analyzed by the total alkali-soluble melanin assay. A melanin standard curve was prepared using a melanin standard and a 1 mol/L NaOH solution. Fiber samples were cut into 1-mm lengths with scissors, then 10 mg of each sample was put into 1 mL of 1 mol/L NaOH in a water bath at 100 °C for 2 h. After the fibers had completely dissolved, 200 μL of the melanin solution was placed into a 96-well plate, and the absorbance value was measured to calculate the melanin content.
Goat skin paraffin sections were first placed in an oven at 65 °C oven to melt the wax, followed by sequential xylene treatment and ethanol dehydration. Sections were then immersed in a ferrous sulfate solution in a 37 °C water bath for 30 min and rinsed five or six times with distilled water. Subsequently, the sections were stained with an acidic potassium ferricyanide solution in a 37 °C water bath for 20 min, followed by three or four rinses with distilled water. Nuclei were counterstained with a solid red solution for 5 min and rinsed three times with distilled water. The sections were then dehydrated through graded ethanol concentrations, cleared with xylene, sealed, and allowed to dry naturally. The fiber length was measured using a ruler, and each sample was measured three times for accuracy.
RNA extraction and purity checking
-
Total RNA was extracted using TRIZOL (TaKaRa, Japan) according to the manufacturer's instructions. The RNA concentration and purity were evaluated using NanoDrop2000 equipment (Thermo Fisher Scientific, USA) with the 260/280 ratios being between 1.8 and 2.0; the 260/230 ratios were greater than 1.6 in all analyzed RNA samples. First-strand cDNA was synthesized using the PrimeScript™ RT reagent Kit (TaKaRa, Japan).
RNA sequencing and data analysis
-
For RNA sequencing, three female goats per breed were selected on the basis of average fiber length. RNA libraries were constructed from 1 μg of total RNA per sample using the Hieff NGS Ultima Dual-mode mRNA Library Prep Kit (Yeasen Biotechnology, China). Poly-T oligo-attached magnetic beads were used to isolate mRNA, which was subsequently reverse-transcribed to synthesize first- and second-strand cDNA. The resulting cDNA was subjected to end blunting, adenylation, and ligation using NEBNext Adaptors. Library fragments were purified with the AMPure XP system, digested with USER Enzyme, and amplified via polymerase chain reaction (PCR) using Phusion High-Fidelity DNA polymerase. The amplified products were purified and quality-checked on an Agilent Bioanalyzer 2100 system. Sequencing was performed on the Illumina NovaSeq 6000 platform to generate 150- bp paired-end reads. Raw data were processed using the online BMKCloud platform. Differentially expressed genes (DEGs) were identified using a threshold of an adjusted p-value [false discovery rate (FDR)] < 0.05 and a |log2 fold change| > 1, and hierarchical clustering of the DEGs was conducted using R software.
Quantitative real-time PCR
-
Total RNA was extracted from the same 12 goat skin samples used for RNA sequencing. Scanned DEGs of melanin deposition and fiber length were based on the Capra hircus genome sequence available from NCBI. NCBP3, SDHA and PTPRA were used as reference genes[25]. Primers were designed using the NCBI website, and the primers are listed in Table 1.
Table 1. Paired primer sequences in qPCR experiments.
Genes Primer sequence (5'–3') Annealing temperature (°C) Amplicon size (bp) ALDOC F: ATTCTGGCCGCAGATGAGTC 60 105 R: GAACAGAACCTGGCGGTACA IRX3 F: AGGGCGGAACAGATCGCT 60 122 R: GAGAGCCGATAAGACCAGAGC NR1D1 F: ACATCGCTGGGAAAGTCAGG 60 102 R: GAGGAAGCCTGGCGTAAACT FOXQ1 F: CGACGGTTGTGGCTTTACTG 60 129 R: TGCTTTCAGGTGGCAGTGAT HOXA9 F: GGAAGAAACGCTGCCCCTAT 60 135 R: TCTTGACCTGCCTCTCCGTA HOXA10 F: CTTCCAAAGGCGAAAACGCA 60 82 R: GTCTGGTGCTTGGTGTAGGG CEBPB F: CCCGCCCGTGGTGTTATT 60 76 R: GGCAGAATGAGAGGCAAGAGT EDA2R F: CTGCTCGTGGTGTTTACCCT 60 118 R: TCTTGCCAGCCTCATACTGC ASIP F: AGCCCAGAGATGAAAGGAACC 60 76 R: GCCACAATAGAGACAGAAGGGA TYR F: CCTCGGCTGATGTGGAGTTT 60 188 R: CTGGGACATCGTTCCGTTCA TYRP1 F: TCAGTTTGTCATCGCCACCA 60 192 R: AGAAATGCTGGTCCCTCGTG DCT F: TTCTCACACCAAGGACCTGC 60 148 R: TGCACACGTCACACTCGTTA PMEL F: GGGCTGACCTTTCCTACACC 60 184 R: ACATGCCTATCTGTGGTGCC SLC24A5 F: TGCACGCTGCAGAAAGATTG 60 103 R: CAAGTGTGCAGTAGCCCAGA SLC45A2 F: CAGATCCTGGTCGGAAGTGG 60 237 R: TGTCTGAGGTTAGGGACCGT HTRA4 F: CGTGGCTTCTGGGGTTTTTG 60 109 R: GGTGACAGGCAGTCCGTTTA NCBP3 F: AGGAACTCCATGAGGGCAGA 60 127 R: GACGTGTGTGCTGACGTTTT SDHA F: CGCTACGACACCAGCTACTT 60 103 R: TGGACCCGTCTTCTATGCAC PTPRA F: AATTCAACGCTCTCCCTGCT 60 131 R: AACTGGTGTCAGATGGACTCG The PCR reaction was performed using the corresponding cDNA as a template in a total volume of 10 µL: 5 µL of TB Green Premix Ex TaqII, 1 µL of cDNA, 0.4 µL of each of the upstream and downstream primers, and 3.2 µL of ddH2O. Thermal cycling was carried out on a Biorad CFX96 Real-Time Fluorescence Quantitation System as follows: Pre-denaturation at 95 °C for 30 s, and annealing at 95 °C for 5 s and 59 °C for 30 s. The cycle threshold (Ct) values were obtained on the basis of the threshold line, which was automatically generated by the quantification system. Relative mRNA expression levels were calculated using the 2-ΔΔCt method with the reference genes as internal controls. The specificity of the cycling reaction was verified by analyzing the melting curve of the quantitative real-time PCR (qPCR) products.
Statistical analysis
-
All statistical analyses were performed using SPSS 18.0 software. Data are expressed as mean ± standard error of the mean (SEM). A p-value < 0.05 was considered statistically significant. Comparisons between two groups were made using the independent-samples t-test.
-
Two goat breeds (DBG and YBG) have solid black coats, whereas BJG and IMCG have white coats with great phenotypic differences. Differences in coat color are mainly influenced by the amount of pigmentation. The results of melanin staining in skin sections showed that melanin was deposited in large quantities in the bulb, trunk, and outer root sheath of the epidermis of black goats, but in small quantities in white goats (Fig. 1). Furthermore, the melanin content in the hair was approximately 22.2 times higher in the black goat group compared with the white goat group (Table 2). The results showed that IMCGs' hair and cashmere lengths were higher than those of the three local Chongqing goat breeds (p < 0.05, Table 2).
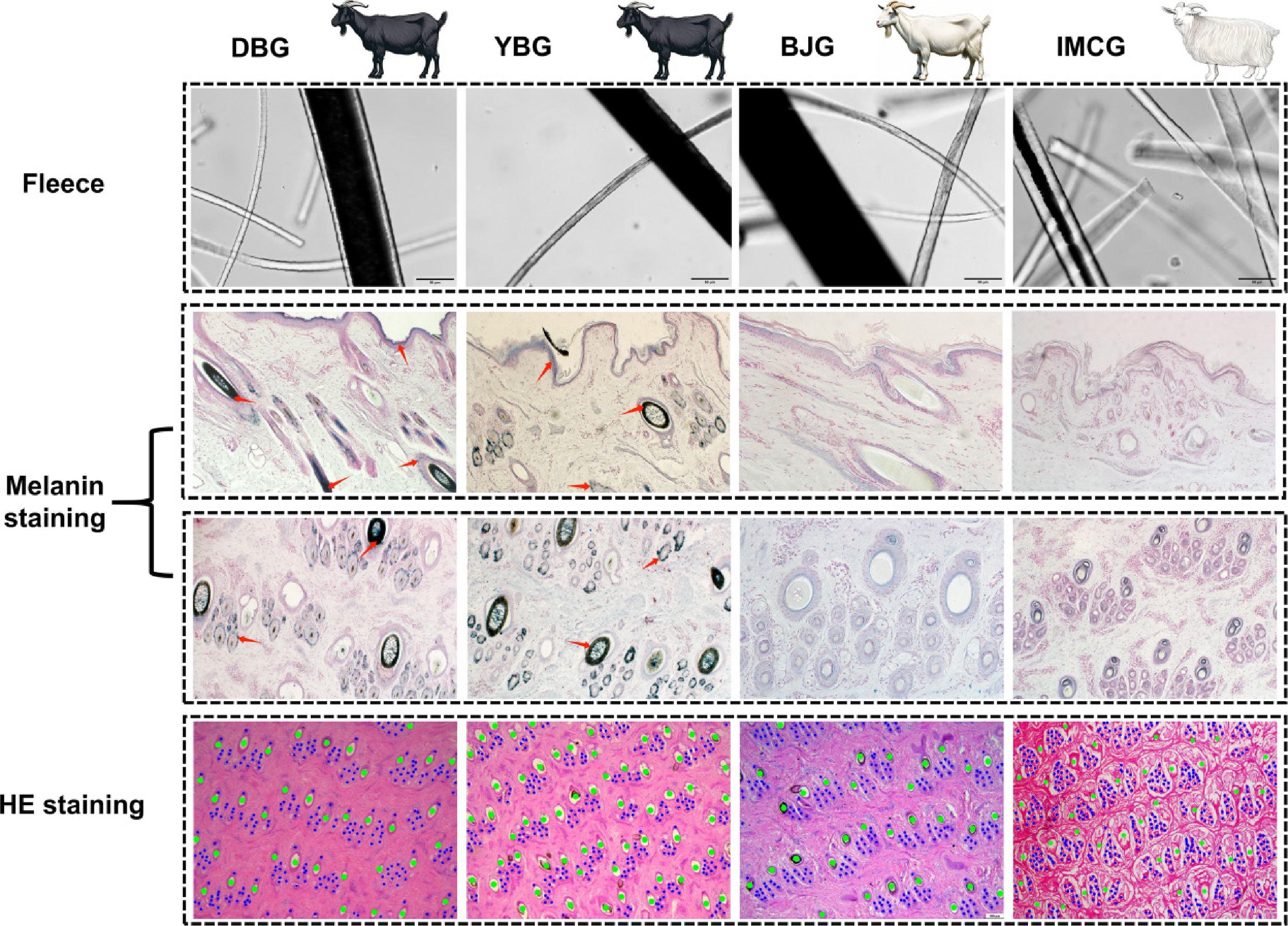
Figure 1.
Histological analysis of four native goat breeds in China. Breeds: Representative animals of DBG, YBG, BJG, and IMCG. Fleece: coarse hair and cashmere under an inverted microscope. Melanin staining: staining results from transverse and longitudinal skin slices of four breeds of goats. The red arrows point to the sites where melanin is deposited.
Table 2. Descriptive statistics of the hair follicle and fiber traits.
Traits Breeds IMCG BJG DBG YBG Fiber traits Hair melanin content (mg) 0.0286 ± 0.0069b 0.0234 ± 0.0135b 0.5509 ± 0.0832a 0.5637 ± 0.0936a Hair length (cm) 11.3 ± 1.6a 4.1 ± 0.6b 3.7 ± 0.6b 4.2 ± 0.8b Cashmere length (cm) 7.4 ± 0.7a 1.3 ± 0.3b 0.4 ± 0.4c 1.2 ± 0.3b Different letters within the same row indicate p < 0.05. Summary of transcriptome data
-
RNA-seq generated a wide range of raw reads in different samples of BJG, DBG, IMCG, and YBG, with a total of 274,187,428 clean reads acquired from 12 sequencing libraries. The alignment of these reads resulted in 96.64%–97.60% of the reads being successfully mapped to the reference genome (ARS1). In addition, all 12 sequenced samples showed a Q30 base percentage above 94.33% and a GC content of ~50%, indicating high-quality RNA-seq data suitable for further analysis. No sequencing bias was detected across the dataset. Detailed statistics for each sample group are presented in Table 3. Gene expression levels of all samples were evaluated using fragments per kilobase per million mapped reads (FPKM). Pearson's correlation coefficients among samples of the same breed were close to 1 (Fig. 2a). High intergroup reproducibility was also confirmed through principal component analysis (PCA) (Fig. 2b). Additionally, the results of the top 2,000 gene clustering results showed consistent expression patterns (Fig. 2c). These results demonstrated strong reproducibility, supporting the reliability of the data for subsequent analyses.
Table 3. Transcriptome sequencing data of 12 skin tissues.
Breed Sample Clean reads Mapping rate (%) Q30 (%) GC content (%) IMCG IMCG-1 21,132,218 96.86 94.34 51.12 IMCG-2 21,693,880 97.41 95.15 50.42 IMCG-3 20,226,767 97.28 94.78 50.21 BJG BJG-1 27,068,839 96.89 94.98 50.92 BJG-2 23,048,523 96.82 95.36 50.73 BJG-3 21,340,575 97.43 94.75 50.10 DBG DBG-1 22,755,384 97.60 95.20 49.35 DBG-2 24,074,908 97.21 94.73 50.36 DBG-3 24,040,968 97.10 95.66 49.97 YBG YBG-1 21,388,934 96.59 94.33 50.86 YBG-2 22,957,714 97.04 95.20 51.48 YBG-3 24,458,718 96.64 94.94 51.14 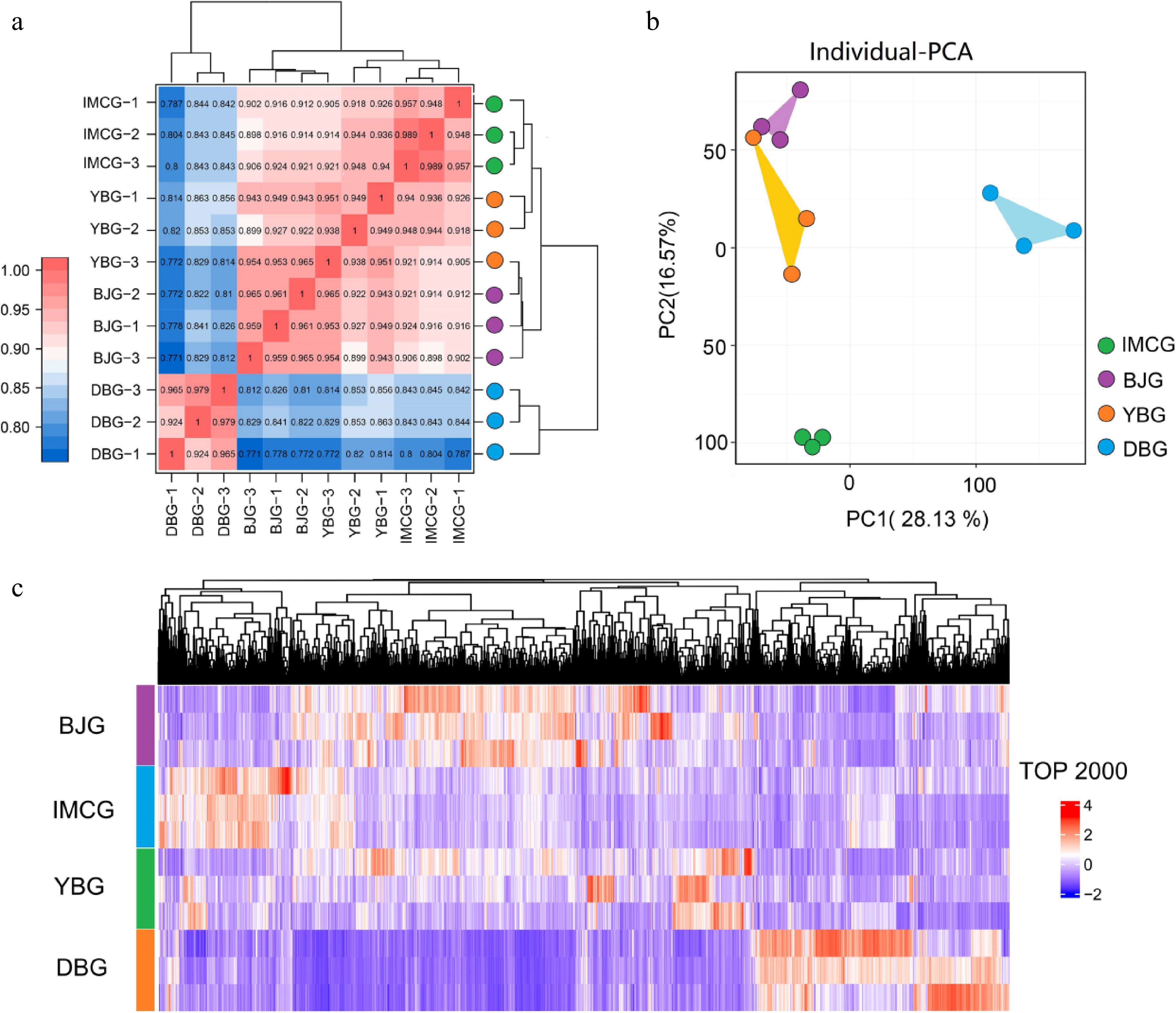
Figure 2.
Transcriptome analysis of skin tissues among four goat breeds.(a) Sample correlation heatmap; (b) PCA (principal component analysis) plot; (c) cluster heatmap of the top 2,000 genes.
DEGs associated with coat color
-
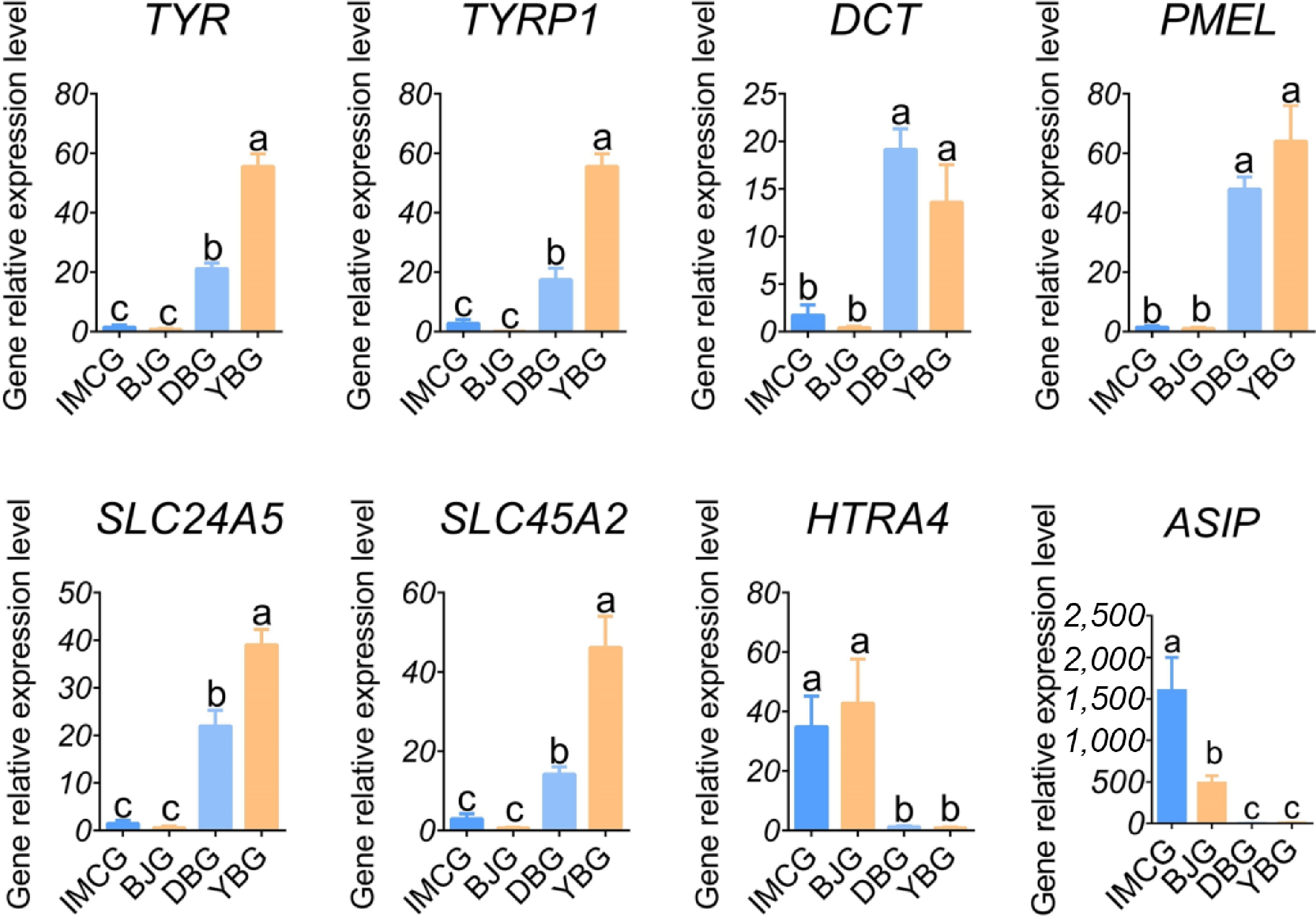
The phenotyping outcomes classified the four breeds in this study into the black goat group (DBG, YBG) and the white goat group (BJG, IMCG). Further statistical analysis of the genes that were differentially expressed by different color groupings was performed to screen candidate key genes for pigmentation of the goat coat. In four black and white goat comparisons, BJG vs. YBG, BJG vs. DBG, DBG vs. IMCG, and YBG vs. IMCG, we identified 252, 2,938, 2,988, and 657 genes, respectively (Fig. 3a). Fifteen overlapping genes were consistently differentially expressed between black- and white-coated goats. Our results showed that TYR, TYRP1, TYRP2, PMEL, SLC24A5, SLC45A2, and UCHL1 were upregulated and the genes ASIP, HTRA4, LAMA2, LOC102178109, NOX4, ATP12A, NOX5, and PRG4 were downregulated (Fig. 3b).
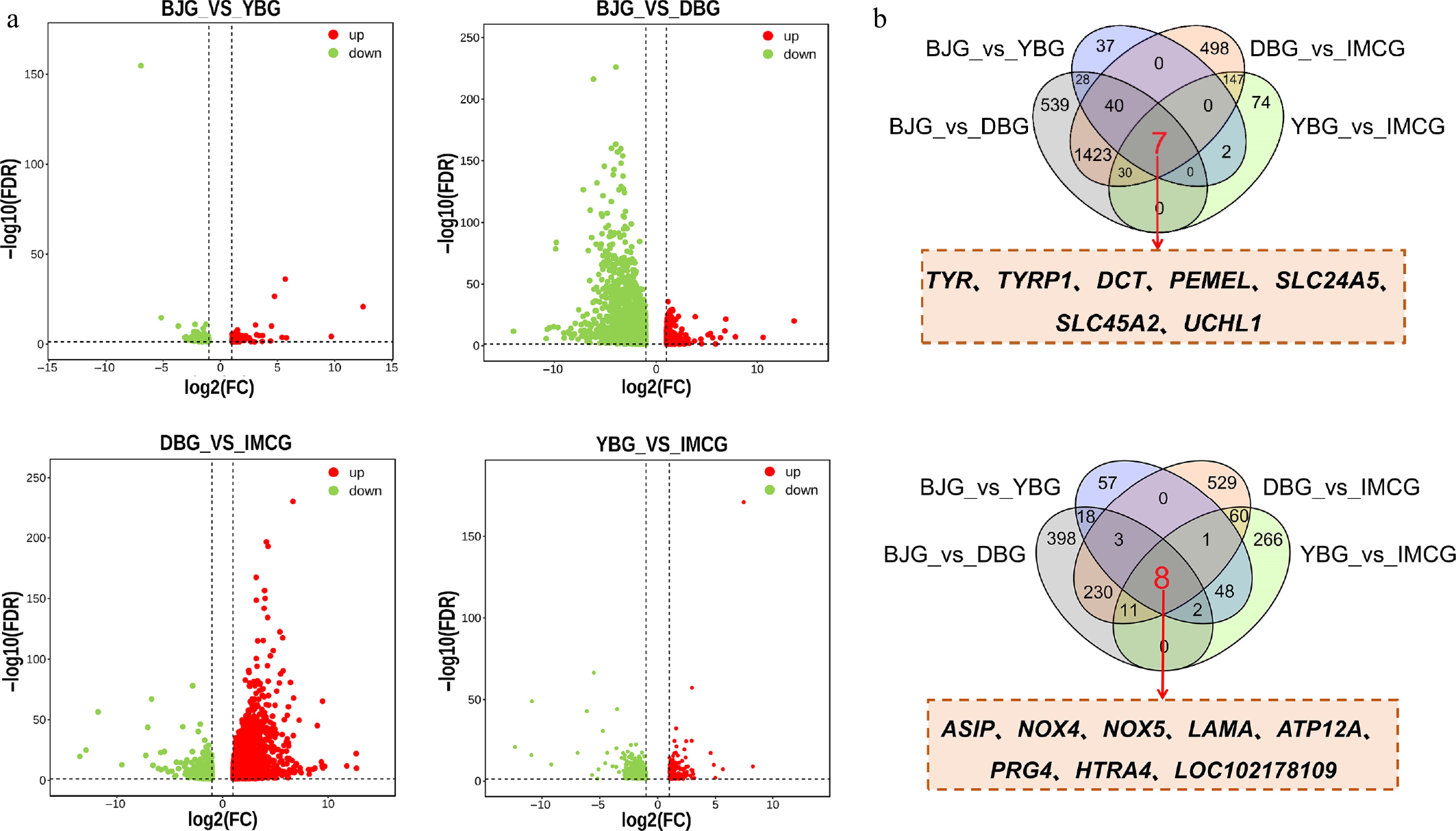
Figure 3.
Identification of core genes related to coat color.(a) Volcano plot of genes among black and white hair goats, including BJG vs YBG, BJG vs DBG, DBG vs IMCG, YBG vs IMCG; (b) overlapping genes among four comparisons.
DEGs associated with fiber length
-
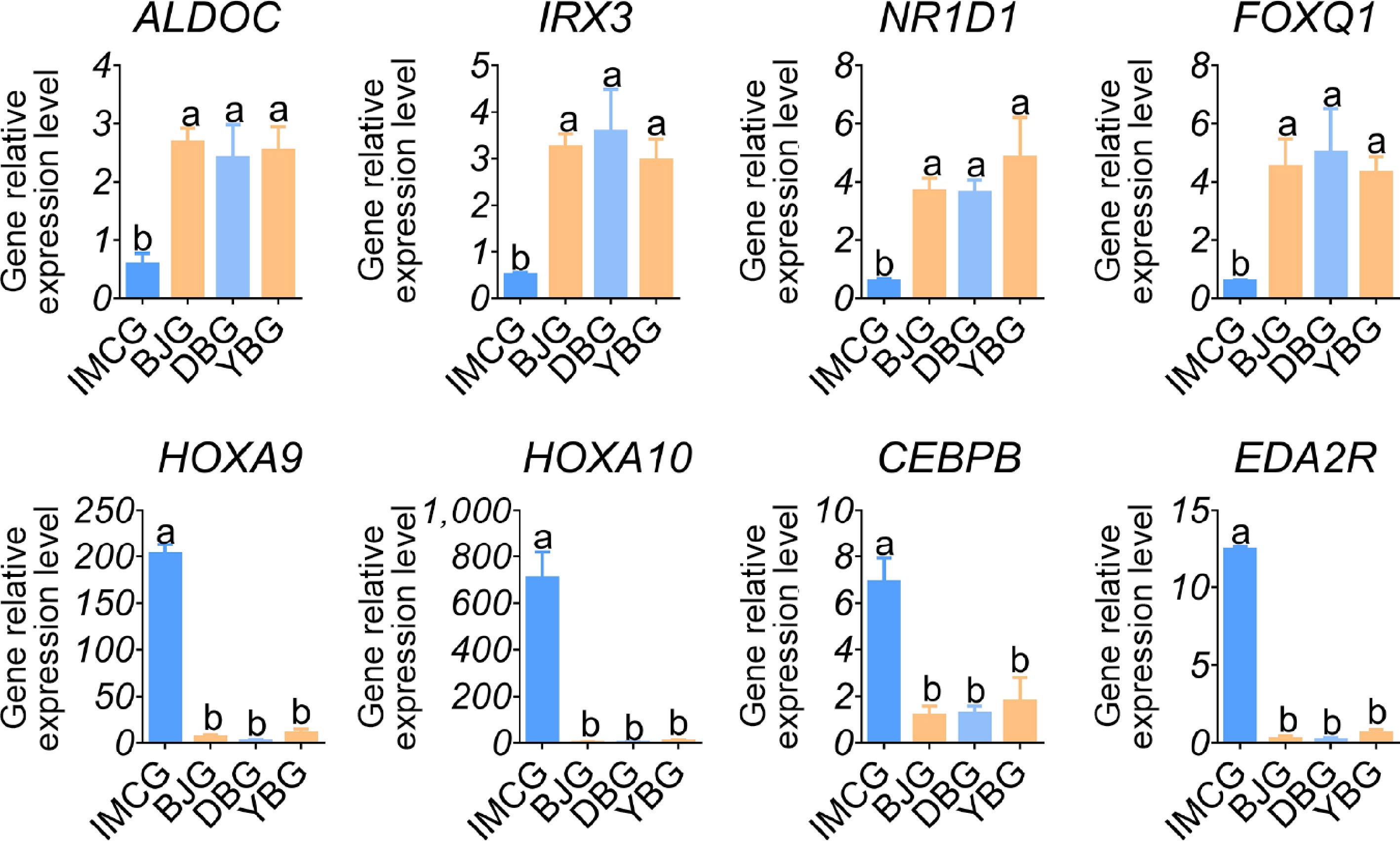
In order to screen the relevant genes regulating fiber length in the IMCG breed, we compared the transcriptome sequencing data of each of the three non-cashmere goat breeds (BJG, DBG, and YBG) with IMCG. Between BJG and IMCG, 1,389 DEGs were identified, with 688 upregulated and 701 downregulated. In the DBG vs. YBG comparison, 2,988 DEGs were identified, including 2,146 upregulated and 842 downregulated genes. In the YBG vs. IMCG comparison, 657 DEGs were identified, including 261 upregulated and 396 downregulated genes (Fig. 4a). Venn analysis was performed to identify stable DEGs and identified 135 co-downregulated and 41 co-upregulated genes. On the basis of these overlapped genes, we performed a literature survey. Genes including FOXQ1, HOXA9, HOXA10, CEBPB, EDA2R, and SOX10 were found that related to 'skin', 'hair follicle', or 'cashmere' (Fig. 4b).
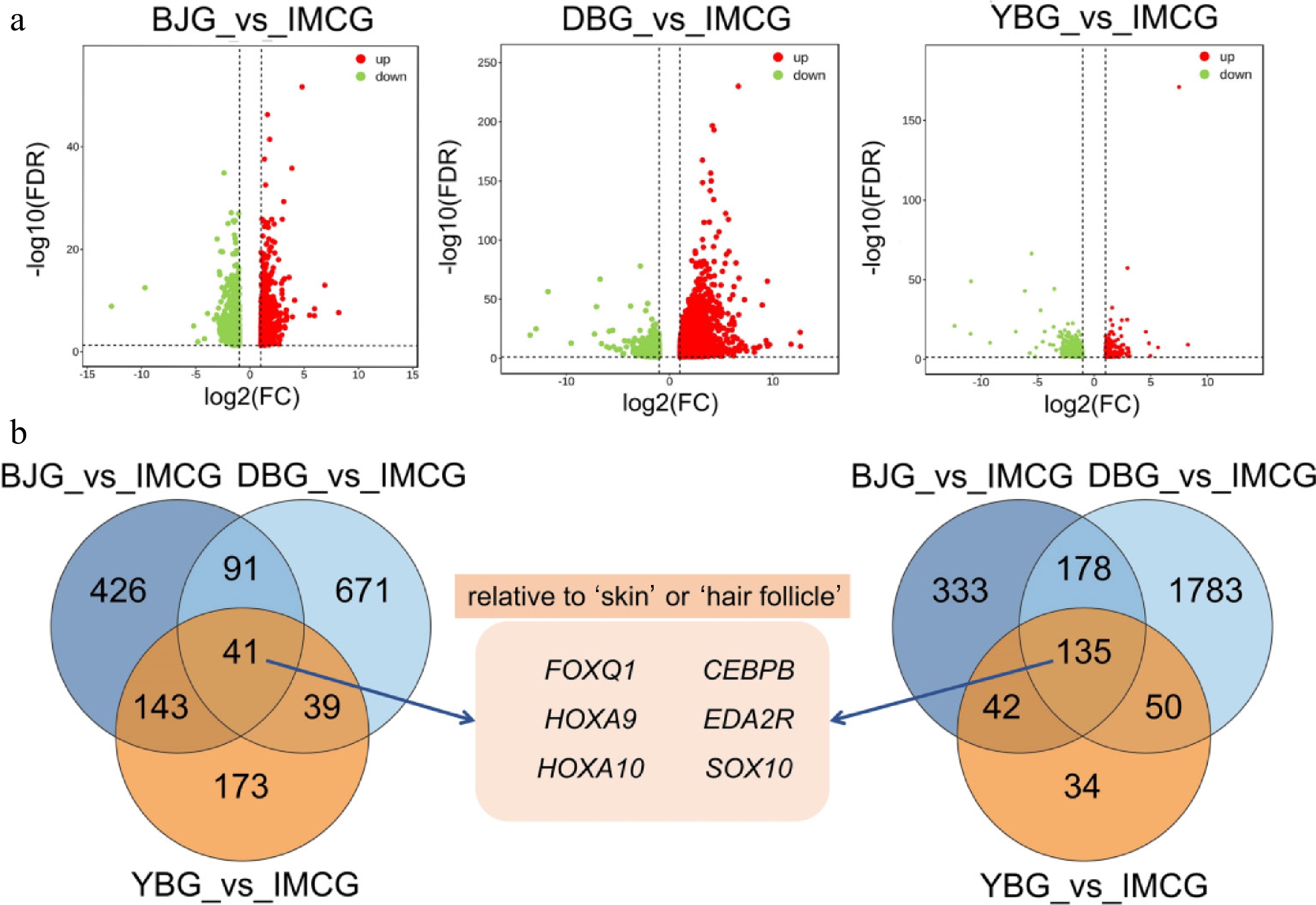
Figure 4.
Identification of core genes related to fiber length. (a) Serial volcano plot of comparisons including BJG vs IMCG, DBG vs IMCG, and YBG vs IMCG. (b) Overlapping genes among three comparisons.
qPCR validation
-
To validate the reliability of the RNA-seq data, we selected 16 overlapping DEGs associated with coat color and fiber length for qPCR analysis. For genes involved in melanogenesis, including ASIP, TYR, TYRP1, TYRP2, PMEL, SLC24A5, and SLC45A2, the qPCR results showed expression trends consistent with the RNA-seq data, confirming the accuracy of sequencing and highlighting their significant roles in regulating pigmentation (Fig. 5). Similarly, the expression patterns of overlapping hair fiber-related genes, ALDOC, IRX3, NR1D1, FOXQ1, HOXA9, HOXA10, CEBPB, and EDA2R, were highly concordant between the qPCR and RNA-seq results, reinforcing their potential involvement in regulating goat hair traits (Fig. 6). Together, these findings validate the reproducibility and reliability of transcriptome profiling.
-
This study investigated the phenotypic and transcriptomic profiles of coat color and fiber length among the four goat breeds. The Venn analysis between any pair of black and white goats revealed seven overlapping upregulated and eight overlapping downregulated genes. The melanogenesis-related genes TYR, TYRP1, DCT, PMEL, SLC24A5, SLC45A2, and ASIP were included, which are relevant to melanin biosynthesis, developmental pigmentation, tyrosine metabolism, and the melanosome function process. Furthermore, in the three cashmere versus non-cashmere goat breeds comparisons, 176 overlapping DEGs were scanned, with 135 genes upregulated and 41 genes downregulated. Several genes, such as FOXQ1, HOXA10, and EDA2R, were identified, which may be associated with fiber length.
Coat color is a key basis for breed identification and strain delineation in goat breeding. With the increasing demand for natural fiber products, coat color has become an important factor influencing the economic value of cashmere goats. Consequently, the breeding of cashmere goats with diverse natural colors has emerged as a new direction in the industry[26,27]. The ASIP gene, a DEG observed in white and black goats, is associated with light coat color and with the regulation of melanin synthesis through the cAMP signal pathway[28]. The transcriptome analysis in Youzhou dark goats and Yudong white goats revealed structural variation in the ASIP gene, explaining its lower expression in the hyperpigmented skin[29]. A previous study reported that a 4-bp deletion in Exon 3 of the ASIP gene was associated with the black coat phenotype in domestic guinea pigs[30]. The tyrosinase protein family members, including tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and dopachrome tautomerase (DCT), are essential factors in the process of melanogenesis, which encode the proteins responsible for synthesizing true and brown melanin. Moreover, they are direct targets of several factors regulating melanin synthesis, functioning as pivotal elements in melanin biosynthesis that determine the quality and quantity of the melanin produced[31]. Knockdown of the 3' untranslated region (UTR) of the rabbit TYR gene by the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing system resulted in a reduction of melanin in the hair follicles and iris and a change in coat color from black to gray[32]. Furthermore, premelanosome protein (PMEL), Solute Carrier Family 24 member 5 (SLC24A5), and Solute Carrier Family 24 member 2 (SLC45A2) are the key structural protein participants in the maturity of melanosomes. PMEL is involved in the maintenance of melanosomes' morphology. Its inactivation results in a change in the shape of melanosomes from oblong to spherical, a reduction in melanin content, and a dilution of the animal's coat color[33,34]. For example, PMEL-/- in mice drastically reduces eumelanin in the hair and dilutes the coat color[35]. Likewise, in goats, high expression of PMEL correlates with a dark coat color[36]. SLC24A5 and SLC45A2 play a key role in melanin synthesis by facilitating the transportation and processing of TYR, TYRP1, and DCT within cells. The significance of those two genes was shown by human and mouse single nucleotide polymorphisms[37,38]. Apar et al. reported that TYRP1, SLC24A4, PMEL, DCT, and ASIP were differentially expressed between black and brown skin tissues[36]. In addition, Wu et al. identified that TYRP1, TYR, DCT, ASIP, PMEL, and ASIP had different expression levels in black and white cashmere goats' skin tissue[39], which is consistent with the results of this study.
Cashmere is an important raw material for the world textile industry and has significant economic value. Among the DEGs observed in cashmere and non-cashmere goat, we confirmed FOXQ1, HOXA10, and EDA2R as genes associated with fiber length. Forkhead box (FOX) proteins have been shown to play important roles in regulating the expression of genes involved in cell growth, proliferation, and differentiation. Some of the Fox family members are important for resting, self-renewal, and differentiation of hair follicle stem cells (HFSCs) throughout the cell cycle. For instance, deletion of FOXE1 results in abnormal hair formation, wherease deletion of FOXI3 disrupts the hair follicle cycle and affects hair regrowth[40]. In addition, a previous study demonstrated that in mice, FOXQ1 mutations produced poorly developed hair shaft medullary cells and influenced hair color and morphology[41]. Moreover, an association between wool fineness and FOXQ1 has been revealed in previous research[42]. A previous study has shown that HOX genes, which are widely expressed in animal skin and hair follicle tissues, play a crucial role in the formation of fetal skin hair follicles and the mature hair follicle cycle[43]. For example, HOXA4, HOXA5, and HOXA6 are expressed in various hair follicle organs during both embryonic and anagen stages. HOXA10 is highly expressed in fetal skin at 10 and 17 weeks, but weakly expressed in neonatal and adult skin, highlighting its role in hair follicle formation[44]. HOXC13 is essential for the hair follicle cycle, whereas mice lacking HOXC13 (HOXC13-/-) display blocked hair follicle genesis and cycling[45]. The injection of recombinant HOXC13 has been shown to prolong the anagen phase, indicating its regulatory role in hair growth and cycling[46]. Specifically, HOXA7 is positively correlated with wool fineness, whereas HOXC13 is linked to wool length[47]. The ectodysplasin A receptor (EDAR) signaling pathway regulates embryonic epidermal cells' fate and hair follicle differentiation, with mutations in EDA or EDAR causing developmental abnormalities in hair follicles[48]. EDA2R, the receptor for EDA-A2, activates the downstream nuclear factor kappa B (NF-κB) pathway when highly expressed alongside EDA-A2; this activation is crucial for hair follicle formation via the WNT/β-catenin pathway[49,50]. Research has demonstrated that EDA-A2/EDA2R signaling plays an inhibitory role in hair growth. Therefore, an inhibitor of this pathway could represent a promising therapeutic agent for the treatment and prevention of hair loss[51]. In goats, EDA2R is also seasonally expressed in cashmere goat skin, with peak levels observed in the late anagen phase, correlating with the suppression of hair growth and the initiation of regression[52]. Collectively, our findings align with and expand upon existing knowledge, emphasizing the roles of transcriptional regulators (FOXQ1, HOXA10) and signaling receptors (EDA2R) in determining fiber length. The consistent differential expression of these genes across breeds highlights their potential as molecular markers for the genetic improvement of the quality and yield of cashmere.
Coat color and fiber length are critical economic traits in goats. The development of naturally colored, high-yielding fiber goats represents a market-driven direction for developing new breeds. Key genes identified through transcriptome sequencing provide a valuable foundation for further goat breeding research. Utilizing gene-editing technologies such as CRISPR/Cas9 to edit these genes could enable targeted modification of specific traits (e.g., coat color and fiber traits) and accelerate the breeding process of superior varieties. For example, on the basis of RNA-seq data, Zhang et al.[53] used CRISPR/Cas9 technology to edit the ASIP gene, generating six ASIP-edited sheep and confirming its influence on wool color. Similarly, Hao et al. edited the EDAR gene and reported that the edited individuals exhibited features such as hairless heads and abnormal skin and follicles[54]. Variations in the FGF5 gene and the introduction of the VEGF gene have also been proven to extend the anagen phase of hair follicles, stimulate hair growth, and thereby enhance cashmere yield and fiber length[55,56]. In this study, several of the identified genes have been confirmed to regulate the relevant traits. However, the remaining candidate genes hold significant potential and warrant further investigation.
-
This study identified a set of core genes (ASIP, TYR, TYRP1, DCT, PMEL, SLC24A5, and SLC45A2) involved in the melanin biosynthesistic pathway in four Chinese native goat breeds. Additionally, FOXQ1, HOXA10, and EDA2R were recognized as candidate genes associated with fiber length. The expression patterns of these genes were validated by qPCR. Overall, this study integrates phenotypic and transcriptomic analysis to reveal key candidate genes influencing coat color and fiber length traits in goats, providing valuable insights for future genetic improvement and breeding strategies.
This work was financially supported by the National Key Research and Development Program of China (No. 2022YFD1300202), Strategic Cooperation Foundation of Chongqing Municipal People’s Government and Chinese Academy of Agricultural Science, Chongqing Modern Agricultural Industry Technology System (CQMAITS202513), and the Collection, Utilization and Innovation of Germplasm Resources by Research Institutes and Enterprises of Chongqing, China (cqnyncw-kqlhtxm).
-
All procedures were reviewed and preapproved by the Animal Experimental Committee of Southwest University, China (IACUC-20230227-01, approval date: 27 February 2023). The research followed the 'replacement, reduction, and refinement' principles to minimize harm to animals. This article provides details on the housing conditions, care, and pain management for the animals, ensuring that the impact on the animals was minimized during the experiment.
-
The authors confirm their contributions to the paper as follows: study conception and design: Zhao Y, Zhang J; data collection: Xiao M, Zhang J, Lv Y, Zhou D, Wang Y, Guo Y; analysis and interpretation of results: Xiao M, Zhang J, Wu Y, Guo J; writing − draft manuscript preparation: Xiao M; writing − review: Zhang J, Zhao Y. All authors reviewed the results and approved the final version of the manuscript.
-
The authors confirm that the original transcriptome sequencing data are deposited with the NCBI Bioproject under accession No. PRJNA1252663.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2026 by the author(s). Published by Maximum Academic Press on behalf of Nanjing Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xiao M, Wu Y, Lv Y, Zhou D, Wang Y, et al. 2026. Integrative skin phenotypic and transcriptomic analyses reveal candidate genes for coat color and fiber length in four Chinese goat breeds (Capra hircus). Animal Advances 3: e009 doi: 10.48130/animadv-0025-0046
Integrative skin phenotypic and transcriptomic analyses reveal candidate genes for coat color and fiber length in four Chinese goat breeds (Capra hircus)
- Received: 08 September 2025
- Revised: 20 October 2025
- Accepted: 30 October 2025
- Published online: 12 February 2026
Abstract: Coat color and fiber length are key traits for breed improvement and fiber production in goats. In this study, fiber samples and skin tissues were collected from four Chinese goat breeds: The solid black group (Dazu black goat and Yudong black goat) and the solid white group (Banjiao goat and Inner Mongolian cashmere goat). Fiber and skin color was evaluated, fiber length was measured, and skin transcriptome sequencing was performed using these samples. Our results showed that skin pigmentation was predominant in the epidermis, hair papilla, hair shaft, and outer root sheath of black goats. Specifically, the melanin content of the black goats' hair was approximately 22.2 times greater than that of the white goats (p < 0.05). Inner Mongolian cashmere goats had longer hair/cashmere length compared with the other breeds (p < 0.05). After reads mapping and gene quantification, differentially expressed genes (DEG) were identified. The Venn analysis between the black and white goats revealed seven overlapping upregulated genes and eight overlapping downregulated genes. Among these, melanogenesis-related genes including TYR, TYRP1, DCT, PMEL, SLC24A5, SLC45A2, and ASIP were validated using quantitative polymerase chain reaction (qPCR). Furthermore, 176 DEGs were identified between cashmere and non-cashmere goat breeds. Among them, fiber length-related genes (FOXQ1, HOXA10, and EDA2R) were differentially expressed, as found by qPCR. In conclusion, the comparative analyses of phenotypes and transcriptomes provide valuable insights into the genetic regulation of coat color and fiber length in goats and establish a foundation for future breeding programs and genetic selection strategies.
-
Key words:
- Goat /
- Coat color /
- Fiber length /
- Skin phenotype /
- Transcriptome