-
Ethylene, a multifunctional phytohormone, regulates diverse biological processes beyond its implications in plant responses to biotic and abiotic stresses, including seed germination, flowering, fruit ripening, senescence, and organ abscission[1]. In climacteric fruits such as tomato, banana, apple, mango, and kiwifruit, ethylene is crucial in ripening, acting not only as an initiator of maturation but also as a pivotal regulator throughout the ripening progression[2]. Current research in tomato and other fruits has elucidated the ethylene biosynthesis, perception, signaling transduction, and downstream gene networks governing ripening processes[3,4]. These studies demonstrate that ethylene biosynthesis is catalyzed by ACC synthase and oxidase, while ethylene perception is initiated through ethylene receptors[5]. The ethylene signal is propagated through positive regulators EIN2/3, culminating in transcriptional activation of downstream ripening-related genes by master regulators ethylene response factors (ERFs), and other transcription factors (TFs)[6,7]. Ethylene-mediated fruit ripening constitutes a complex yet precisely orchestrated biological process, characterized by coordinated changes in pigmentation, aroma profile, flavor compounds, and textural properties, all driven by selective activation of ripening-specific functional genes[7]. TFs orchestrate spatiotemporal gene expression patterns through sequence-specific binding to cis-regulatory elements in promoter regions, directing transcriptional activation or repression of target genes with precise quantitative, temporal, and spatial control[8−10]. The discovery and in-depth characterization of tomato ripening mutants rin, nor, and cnr have highlighted the pivotal role of TFs in governing fruit ripening, drawing significant scientific attention to their regulatory mechanisms[11]. The proteins encoded by the genes RIN, NOR, and CNR are members of the MADS-box, NAC, and SBP families of TFs, respectively[12−14]. NAC (NAM, ATAF1/2, CUC2) TFs are among the largest plant-specific TF families, orchestrating pivotal regulatory functions in the ripening of diverse fruits[15]. For example, the tomato TFs NOR and its homolog Nor-like directly modulate genes that contribute to ethylene biosynthesis, carotenoid accumulation, and cell wall degradation to coordinate fruit ripening and quality formation[16]. In litchi, LcNAC002 has been identified as a positive factor that coordinates chlorophyll degradation and anthocyanin biosynthesis by directly binding to LcSGR, LcPAO, and LcMYB1 promoters to induce synchronized chlorophyll degreening and red pigmentation[17]. The ClNOR drives a transcriptional cascade in watermelon fruit by activating the TF ClbZIP1, which upregulates ABA biosynthesis and chromoplast development genes, enhancing ABA accumulation and flesh color development during fruit ripening[18]. These studies demonstrate that NACs play a pivotal regulatory role in orchestrating hormonally controlled fruit ripening and quality dynamics.
TFs orchestrate gene regulation not only individually but also via cooperative or competitive complex formation, dynamically rewiring transcriptional networks[19,20]. The peach TF PpJAM2/3 antagonizes PpMYC2 by competitively binding to identical promoter elements, suppressing transcriptional activation of lignin biosynthetic genes in fruit[21]. Conversely, apple MdCPCL-MdILR3L interaction synergistically upregulates MdGLDH and MdANS expression, enhancing ascorbic acid and anthocyanin accumulation[22]. Beyond protein-protein interactions (PPIs), TFs can also form transcriptional cascades that regulate fruit ripening and quality traits. Illustratively, MdABI5 initiates a transcriptional cascade by activating MdMYBS1, which directly upregulates carotenoid genes (MdPSY2-1 and MdLCYb), and ABA synthesis gene (MdNCED1), driving fruit color development in apple[23]. The latest research found that MdWRKY71 not only regulates the expression of the MdMADS1 gene but also interacts with the MdMADS1 protein to synergistically enhance their activation of downstream anthocyanin biosynthesis genes, thereby mediating ALA-induced anthocyanin accumulation[24]. Therefore, elucidating the regulatory interplay among ripening-associated TFs is fundamental to systematically assembling the transcriptional network governing fruit ripening.
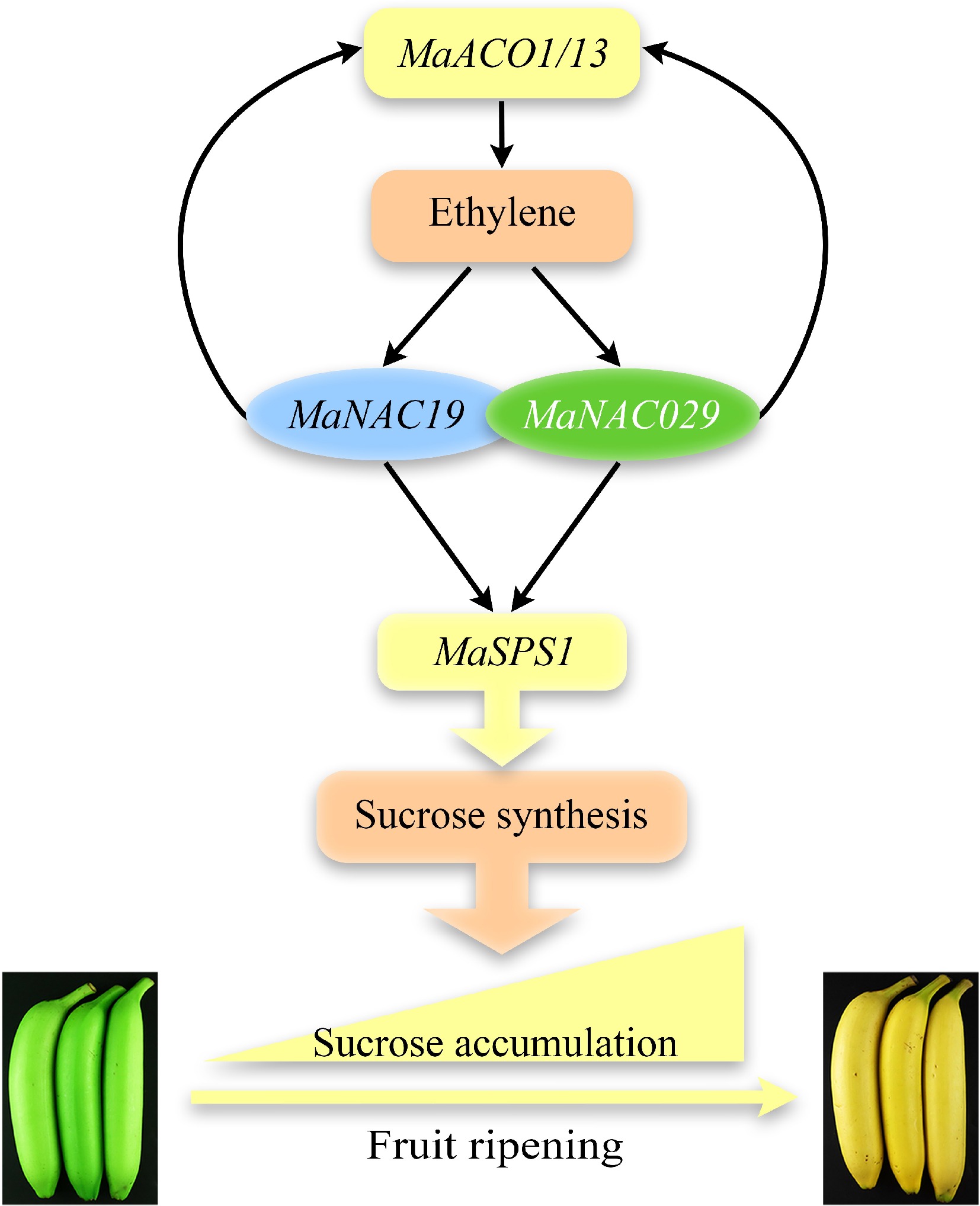
Banana is a globally significant cash crop and the fourth staple food crop in developing countries, as designated by the Food and Agriculture Organization, with its trade volume and value consistently ranking among the highest for fresh fruits worldwide[25]. As a classic climacteric fruit exhibiting high ethylene sensitivity, banana undergoes ethylene-triggered rapid ripening during ambient storage and transport, significantly limiting postharvest shelf life[26]. To enable extended storage and distribution, commercial harvesting occurs at the physiologically mature green stage, followed by targeted ethylene-induced ripening in consumer markets to achieve optimal edibility[27]. Under ethylene action, banana undergoes a series of physiological and biochemical changes, including peel yellowing, aroma release, starch degradation, sucrose accumulation, and fruit softening, thereby developing their characteristic fruit quality attributes[28]. Our recent study revealed that MaNAC029 regulates ethylene biosynthesis throughout banana ripening by directly targeting ethylene synthesis genes MaACO1/13[29]. Concurrently, another NAC TF, MaNAC19, promotes sucrose synthesis via transcriptional activation of the sucrose phosphate synthase gene MaSPS1[30]. The functional interplay between MaNAC029/19, however, has not been elucidated. In this study, it was found that MaNAC029 and MaNAC19 exhibit synchronized expression dynamics during banana fruit ripening. We further demonstrated direct PPI between MaNAC029 and MaNAC19. Critically, MaNAC029 binds to the MaSPS1 promoter, while MaNAC19 occupies promoters of MaACO1/13, demonstrating reciprocal targeting of each other's downstream genes. Notably, the MaNAC029-MaNAC19 interaction synergistically enhances transcriptional activation of MaACO1/13 and MaSPS1. Collectively, these results establish MaNAC029-MaNAC19 as a cooperative transcriptional module that integrates ethylene and sucrose biosynthesis pathways, expanding our comprehension of transcriptional regulation of banana ripening and quality formation.
-
Banana (Musa acuminata, AAA group, cv. Cavendish) fruits at 75%–80% maturation were sourced from a plantation near Guangzhou, China, and treated with 100 μL/L ethylene[29]. The Color Index (CI), ethylene production, firmness, and sucrose content were assessed per established protocols[30]. The specimens were flash-frozen in liquid nitrogen and maintained at –80 °C until utilization.
Gene expression analysis
-
The extraction of total RNA from fruit was performed utilizing the hot borate technique. The qRT-PCR reactions were conducted utilizing the CFX96 Real-Time PCR (Bio-Rad) and the Hieff® qPCR SYBR Green Kit (Yeasen). The design of primer sequences was carried out using Primer software (v5.0), using MaRPS4 as the internal reference gene.
Yeast two-hybrid (Y2H) assay
-
Y2H assays were conducted using the Clontech Matchmaker™ Gold Yeast Two-Hybrid System. Full-length of MaNAC029/19 exhibited auto-activation, whereas their N-terminal regions did not[29,30]; therefore, full-length constructs were cloned into the AD vector, and N-terminal fragments were inserted into the BD vector. The resulting fusion plasmids were co-transformed into Y2HGold cells using the lithium acetate method. The possible PPIs were assessed based on cell growth on selective media and α-galactosidase activity.
Bimolecular Fluorescence Complementation (BiFC) assay
-
MaNAC029/19 full-length subcloned into the pUC-pSPYNE or pUC-pSPYCE vectors, respectively, using NLS-mCherry as the nuclear marker. The resultant vectors, as well as empty plasmids and NLS-mCherry, were co-transfected into BY-2 protoplasts by polyethylene glycol methods[29]. YFP and mCherry fluorescence signals were captured using a Zeiss Axioskop 2 plus fluorescence microscope.
Co-immunoprecipitation (Co-IP) assay
-
MaNAC029/19 full-lengths were cloned into the pEAQ-His and pEAQ-GFP vectors, respectively[31]. MaNAC029-His and MaNAC19-GFP or empty GFP proteins were subjected to transient expression in tobacco leaf tissue by Agrobacterium-modulated transfection. Protein complexes were immunoprecipitated using an anti-GFP antibody (Abcam) and subsequently analyzed by western blotting with anti-His (Abcam) and anti-GFP antibodies, respectively.
Electrophoretic mobility shift assay (EMSA)
-
GST-tagged MaNAC029/19 were purified and obtained in our previous study[29,30]. The fragment containing the NAC TF recognition sequence (NACRS) in MaACO1/13 and MaSPS1 promoters was synthesized and labeled with biotin at the 5' end. EMSA was carried out through a commercialized EMSA kit (Thermo Scientific). Biotin-labelled probes were incubated with GST-MaNAC029 or -MaNAC19 recombinant protein in binding buffer. The free and bound probes were subjected to separation on an acrylamide gel. Unlabelled and mutant probes were utilized as competitors while using the GST protein as a negative control.
Dual-luciferase reporter (DLR) transient expression assay
-
MaACO1/13 and MaSPS1 gene promoters were inserted into the pGreenII 0800-LUC double reporter vector[32] to fuse them with the Firefly luciferase (LUC) reporter gene, respectively. In parallel, MaNAC029/19 full-length was ligated into the pGreenII 62-SK vector[32] to serve as the effector, respectively. To test whether MaNAC029 induces MaSPS1 promoter activity, the single effector (MaNAC029) and the reporter plasmid (CaMV35S-REN/MaSPS1 pro-LUC) were co-transferred into tobacco leaves via Agrobacterium-modulated transient transformation. Analogous to this, the single MaNAC19 effector and the CaMV35S-REN/MaACO1 pro-LUC or CaMV35S-REN/MaACO13 pro-LUC reporter plasmids were co-transformed into tobacco leaves to test whether MaNAC19 induces the promoter activity of the ethylene synthesis genes. To determine the regulatory effects of cooperative MaNAC029 and MaNAC19 on the three promoters, two types of effector plasmids (MaNAC029/19) and the reporter plasmid were co-transfected into tobacco leaves. LUC and REN activities were quantified 60 h after infiltration using a commercial DLR assay system (Yeasen). The target promoter transcriptional activation was indicated by the ratio of LUC to REN. At least six transient assay measurements were included for each pair.
Statistical analysis
-
Data analysis utilized SPSS v19.0. Values represent mean ± SE from three or six independent replicates. Differences among treatment groups were determined by Student’s t-test or Tukey test, as appropriate.
Primers
-
Supplementary Table S1 lists primers employed in this study.
-
To investigate the potential association between MaNAC029 and MaNAC19, their gene expression patterns were analyzed during banana postharvest ripening. Following ripening induction by exogenous ethylene, peel color transitioned from green to yellow by day 3, developing full yellow pigmentation by day 5 (Fig. 1a). This color evolution was quantitated by a rapid increase in the CI from –8.728 at day 0 to 0.296 at day 7 (Fig. 1b). Endogenous ethylene production increased significantly by day 1, peaked on day 3, and subsequently declined (Fig. 1c). Pulp firmness began decreasing on day 3, reaching a minimum value of 7.412 N by day 7 (Fig. 1d). Conversely, sucrose content increased progressively from 19.870 mg/g at day 0 to 80.837 mg/g at day 7 (Fig. 1e). Notably, the expression profiles of MaNAC029 and MaNAC19 exhibited parallel trends, with both genes increasing significantly during ripening, peaking synchronously on day 3, and gradually declining thereafter (Fig. 1f), mirroring ethylene production dynamics (Fig. 1c). These results suggest that MaNAC029 and MaNAC19 may serve as synergistic co-regulators of banana fruit ripening and the formation of quality attributes.
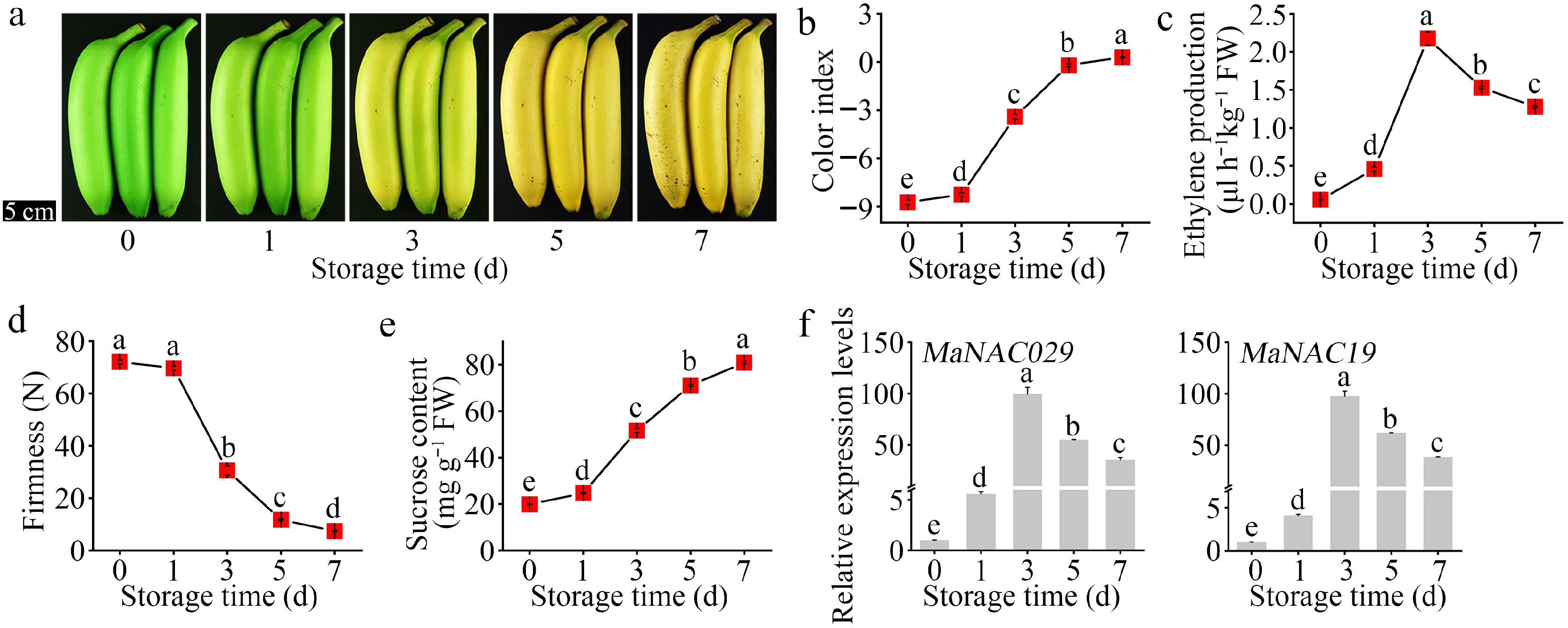
Figure 1.
Expression of MaNAC029 and MaNAC19 during the banana fruit ripening process. (a) Appearance of banana fruit during ethylene-induced ripening. Changes in (b) color index, (c) ethylene production, (d) fruit firmness, and (e) sucrose content during fruit ripening. (f) Expression of MaNAC029 and MaNAC19 in banana fruit during ripening process. The relative expression level is shown as a ratio relative to that on day 0, which was set at 1. Error bars in (b)−(e), and (f) represent SE from six and three replicates, respectively (p < 0.05, Tukey test).
MaNAC029 physical interaction with MaNAC19
-
Given the comparable expression patterns of MaNAC029 and MaNAC19, it is plausible that these proteins interact. Accordingly, we first performed a yeast two-hybrid assay to test this interaction. Due to auto-activation by full-length MaNAC029 and MaNAC19 in yeast cells[29,30], auto-activation-deficient N-terminal truncations (MaNAC029-N and MaNAC19-N) were employed as baits in vectors BD-MaNAC029-N and BD-MaNAC19-N. Full-length counterparts served as prey in AD-MaNAC029 and AD-MaNAC19 vectors. Yeast cells co-transformed with BD-MaNAC029-N and AD-MaNAC19, BD-MaNAC19-N and AD-MaNAC029, as well as the positive control yeast cells that contained p53-BD and T-antigen-AD could grow on selective medium that lack Leu, Trp, His, and Ade, and turned blue in chromogenic substrate α-Gal presence (Fig. 2a). While negative control combinations (BD-MaNAC029-N/MaNAC19-N and empty AD, empty BD and AD-MaNAC029/MaNAC19, and BD-Lamin and AD-T-antigen) fail to cause the growth of transformed yeast cells (Fig. 2a). These results indicated the interaction between MaNAC029 and MaNAC19 in yeast cells.
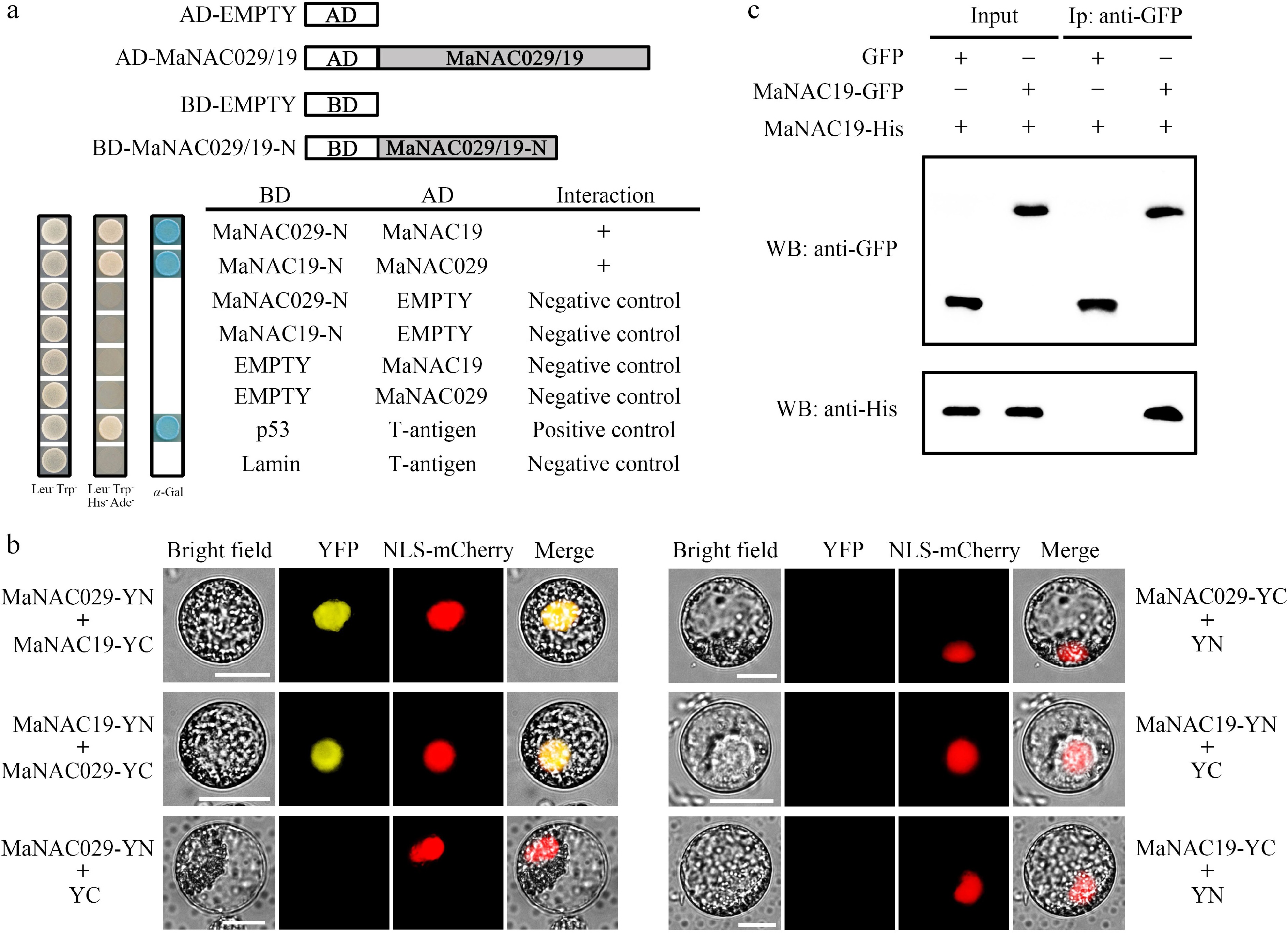
Figure 2.
MaNAC029 interacts with MaNAC19. (a) Y2H assay for the interaction between MaNAC029 and MaNAC19. Yeast cells co-transformed with bait vector and prey vector were grown on selective medium (lacking tryptophan, leucine, histidine, and adenine), and were then stained by X-α-gal. (b) BiFC analysis of the interaction between MaNAC029 and MaNAC19 in tobacco BY-2 protoplasts. MaNAC029/19 were fused with the C terminus of YFP (YC) and the N terminus of YFP (YN), respectively, as indicated, and co-transfected into protoplasts. NLS-mCherry was used as a nuclear marker. Bars, 25 μm. (c) Co-IP assays of MaNAC029-MaNAC19 interaction. MaNAC19-GFP and MaNAC029-His, or empty GFP and MaNAC029-His, were transiently expressed in tobacco leaves and immunoprecipitated with anti-GFP antibody. Immunoprecipitated samples and input controls were detected with anti-GFP and anti-His antibodies, respectively.
To further validate this interaction in planta, BiFC and Co-IP assays were performed. The BiFC assay showcased restored YFP fluorescence in tobacco BY2 protoplasts co-expressing MaNAC029-YN/MaNAC19-YC or MaNAC19-YN/MaNAC029-YC (Fig. 2b). The fluorescence signal co-localized with the nuclear marker NLS-mCherry. By contrast, there was no signal in the negative controls co-expressing MaNAC029/MaNAC19-YN with empty YC vector, or MaNAC029/MaNAC19-YC with empty YN vector (Fig. 2b). These results demonstrate that MaNAC029 interacts with MaNAC19 in the nucleus. For Co-IP assays, protein extracts expressing MaNAC19-GFP or control GFP were immunoprecipitated with anti-GFP conjugated agarose beads. Subsequent immunoblot analysis specifically detected MaNAC029-His only in the MaNAC19-GFP co-expression samples, with no signal observed in GFP-only controls (Fig. 2c). Collectively, these results demonstrate a physical interaction between MaNAC029 and MaNAC19 in living plant cells.
MaNAC029 binds to the MaSPS1 promoter and activates its expression
-
Our previous study identified MaNAC19 as an upstream transcriptional regulator of MaSPS1, a sucrose phosphate synthase gene responsible for the conversion of starch into sucrose in banana fruit during the ripening process[30]. Given the demonstrated MaNAC029-MaNAC19 interaction (Fig. 2), we infer that MaNAC029 may also regulate MaSPS1. We therefore performed in vitro EMSA to assess direct binding of MaNAC029 to the MaSPS1 promoter. Figure 3a illustrates that the purified GST-MaNAC029, but not GST alone, was able to bind directly to DNA fragments that contain the NACRS derived from the MaSPS1 promoter, resulting in obvious mobility shifts. Moreover, the shifting of bands was significantly reduced by adding increasing amounts of unlabeled probes with the same sequence, but not by the mutated competitors (Fig. 3a). Altogether, MaNAC029 specifically targets the MaSPS1 promoter.
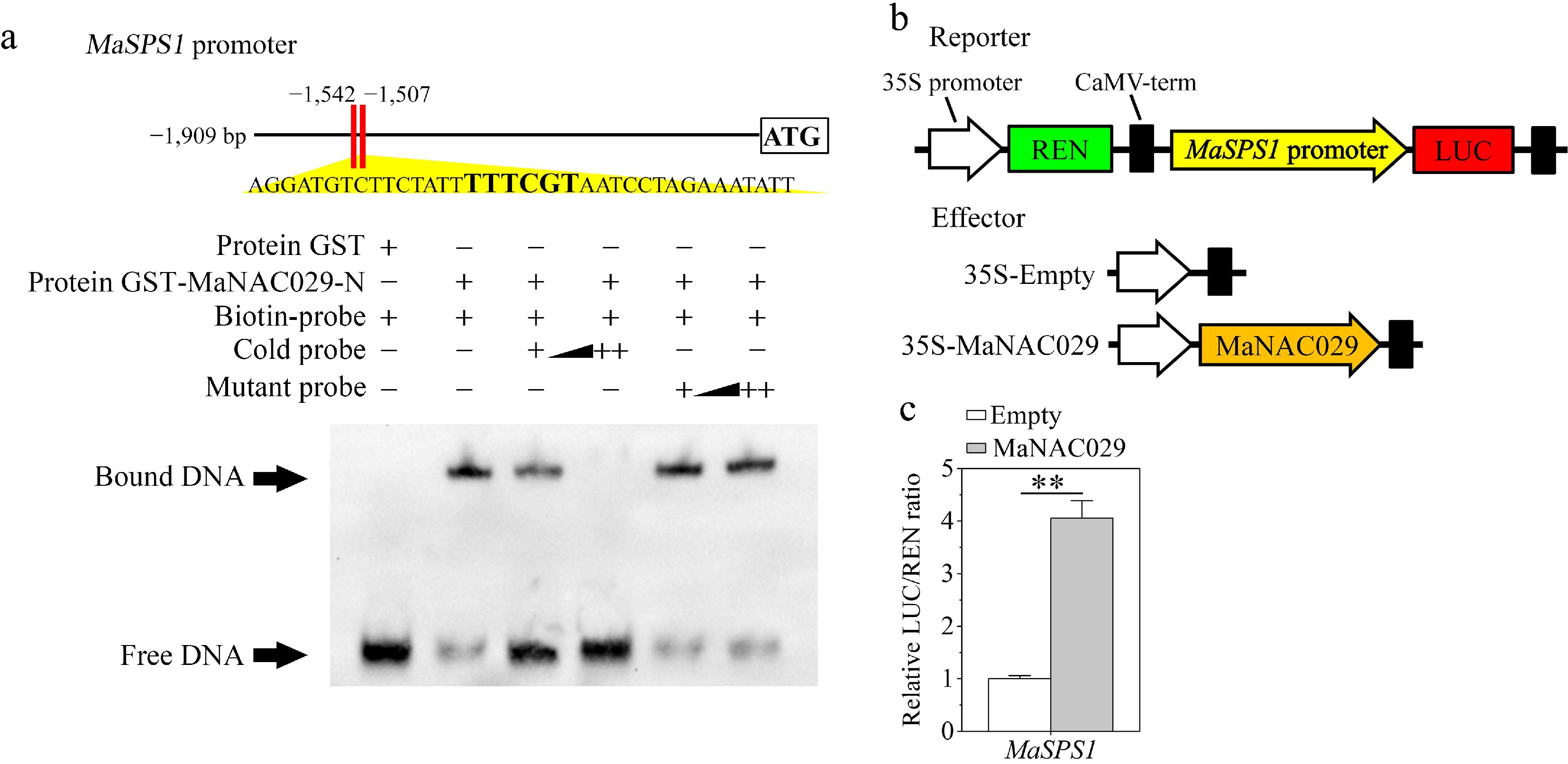
Figure 3.
MaNAC029 induces the expression of MaSPS1 by binding directly to its promoter. (a) EMSA demonstrates the in vitro association of MaNAC029 with the MaSPS1 promoter. A schematic illustration of the MaSPS1 promoter probe is displayed on top, with bold letters denoting NACRS. The negative control, which was the purified GST protein or the recombinant GST-MaNAC029 protein, was subjected to an incubation process along with the probe. Triangles show competing quantities of unlabeled wild-type and mutant probes. MaNAC029 enhances MaSPS1 promoter activity. (b) Schematic representation of reporter and effector constructs. (c) MaNAC029 enhances the MaSPS1 promoter activity in a dual luciferase assay in tobacco leaves. LUC to REN ratio of an empty vector with the MaSPS1 promoter was utilized as a calibrator (deemed 1). Error bars represent SE from six replicates (Student's t-test, ** p < 0.01).
Subsequently, a transient DLR assay was conducted in tobacco leaves to investigate the regulatory impact of MaNAC029 on MaSPS1 transcription. The MaSPS1 promoter was fused with the LUC reporter, and the 35S promoter-driven REN reporter gene was employed as an internal control within the identical vector (Fig. 3b). The effector construct is the 35S promoter-driven MaNAC029 cDNA (Fig. 3b). The LUC/REN ratio was significantly elevated when the MaSPS1 pro-LUC reporter construct was co-transfected with 35S pro-MaNAC029, unlike the empty construct-co-transfected control (Fig. 3c), implying that MaNAC029 activated MaSPS1 promoter activity.
Collectively, MaNAC029 binds to the MaSPS1 promoter and activates its expression.
MaNAC19 directly induces MaACO1/13 transcription
-
Previous studies have revealed that MaNAC029 directly regulates key ethylene biosynthesis genes MaACO1/13[29]. Building upon the interaction between MaNAC029 and MaNAC19 identified in this study (Fig. 2), it is hypothesized that MaNAC19 may also regulate MaACO1/13. This speculation was validated through in vitro EMSA and in vivo DLR transient expression assays. EMSA showed specific binding of purified GST-MaNAC19 to NAC recognition sites (NACRs) in the promoters of MaACO1/13, inducing distinct mobility shifts (Fig. 4a). Unlabeled wild-type probes dose-dependently abolished these shifts but were unaffected by mutated competitors. The DLR transient expression assays demonstrated a marked enhancement in the promoter activities of MaACO1/13 by MaNAC19, yielding significantly elevated LUC/REN ratios relative to the control (Fig. 4b). Together, MaNAC19 directly binds to MaACO1/13 promoters, functioning as a transcriptional activator of these ethylene biosynthetic genes.
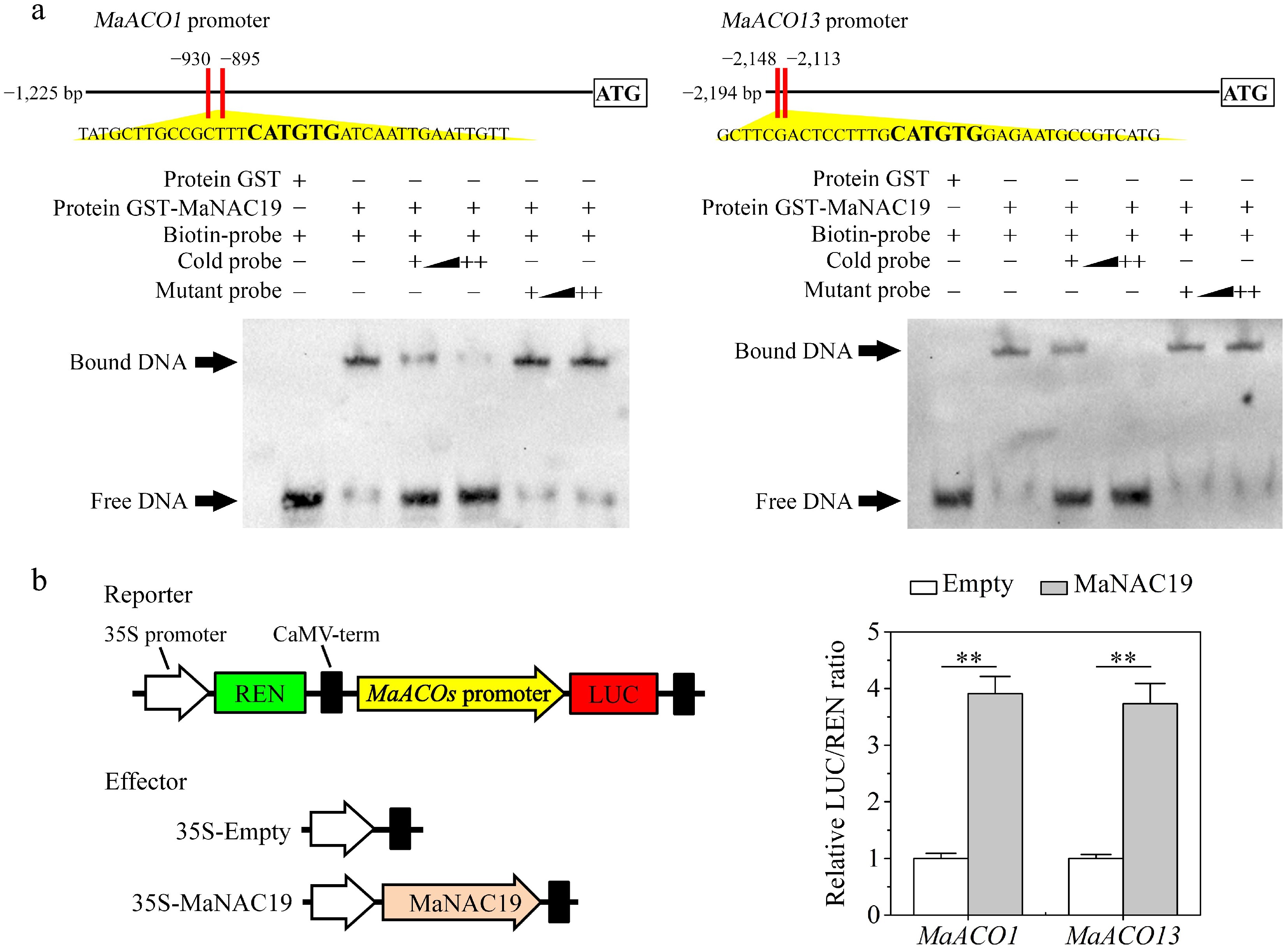
Figure 4.
MaNAC19 directly binds to the promoters of MaACO1 and MaACO13 and enhances their activities. (a) EMSA assay for MaNAC19 binding to MaACO1/13 promoters. The probe sequences corresponding to promoters of ethylene biosynthesis genes are shown at the top of the image, with bold letters representing the NACRs. The purified GST (negative control) or recombinant GST-MaNAC19 protein was subjected to an incubation process along with probes. Triangles indicate increasing amounts of unlabeled wild-type or mutated probes for competition. (b) MaNAC19 enhances the promoter activity of MaACO1/13 in a DLR assay in tobacco leaves. Schematic representation of reporter and effector constructs is shown in the left panel. The value of LUC/REN of the empty vector plus the promoter reporter was used as a calibrator (set as 1). Error bars represent SE from six replicates (Student's t-test, ** p < 0.01).
MaNAC029 and MaNAC19 synergistically stimulate promoters of genes governing ethylene biosynthesis and sucrose accumulation
-
Given that MaNAC029 and MaNAC19 interact with each other and share target genes, DLR assays were next performed in tobacco leaves to ascertain whether MaNAC029 and MaNAC19 could impact each other's action in trans-activating the expression of sucrose phosphate synthase and ethylene biosynthesis genes (Fig. 5a). Unlike, co-transfection with the empty effector, MaACO1/13 or MaSPS1 promoter co-transfection with MaNAC029 or MaNAC19 significantly enhanced promoter activity with a relatively higher LUC/REN ratio (Fig. 5b). Critically, combined expression of both TFs synergistically amplified transcriptional activation across MaACO1/13 and MaSPS1 gene promoters, exceeding levels achieved by individual MaNAC029 or MaNAC19 expression (Fig. 5b). These results establish MaNAC029-MaNAC19 as a cooperative regulatory module that synergistically transactivates sucrose phosphate synthase and ethylene biosynthesis genes.
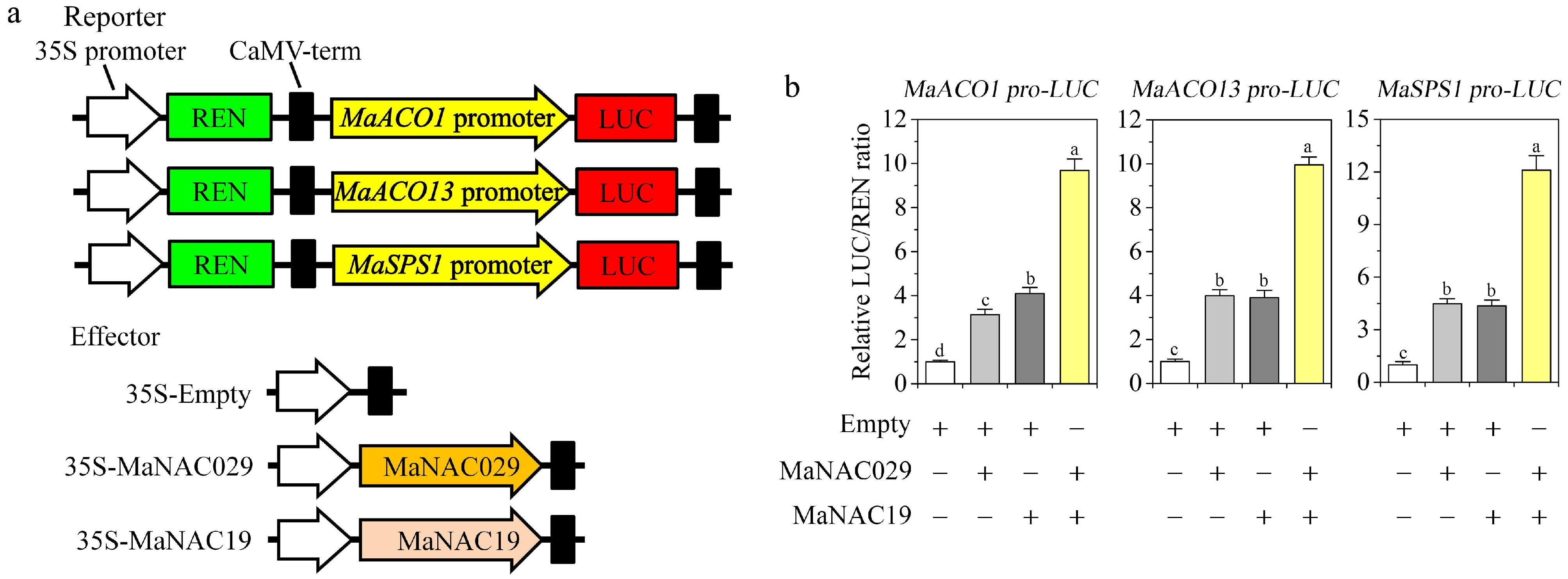
Figure 5.
MaNAC029 and MaNAC19 synergistically trans-activate the ethylene biosynthesis genes (MaACO1/13) and sucrose phosphate synthase gene (MaSPS1) in tobacco leaf dual-luciferase reporter (DLR) assays. (a) Schematic of reporter and effector constructs. (b) Activation ofMaACO1/13 and MaSPS1 promoters by individual or combined expression of MaNAC029 and MaNAC19. LUC/REN ratios of the empty vector control were set to 1.0 for normalization. Data: mean ± SE (n = 6 replicates). Different letters above the bars indicate significant differences (p < 0.05, Tukey test).
-
At the mature green pre-climacteric stage, bananas are harvested and require ethylene-induced ripening after storage and transport to develop good edible quality[27]. Therefore, ethylene-induced postharvest ripening is the critical stage determining the market value of commercial bananas. Postharvest ripening and quality formation in banana fruit result from the selective expression of multifunctional genes orchestrated by an extensive network of TFs. Our recent studies identified MaNAC029/19 as upstream TFs that directly activate ethylene biosynthesis genes and sucrose synthesis genes, respectively, during banana ripening[29,30]; however, their functional crosstalk and regulatory hierarchy remain unelucidated. Herein, we identified a PPI between MaNAC029/19 and discussed the molecular mechanism by which the MaNAC029-MaNAC19 transcriptional module regulates banana fruit ripening and quality formation.
As a climacteric fruit, bananas rely critically on ethylene during ripening, making the regulatory mechanisms of ethylene biosynthesis a sustained research focus in postharvest biology of banana fruit[26]. Our recent study reveals that MaNAC029 promotes ethylene biosynthesis during banana ripening by directly activating the MaACO1/13 genes transcription[29]. The present study further reveal that MaNAC029 and MaNAC19 exhibit identical expression patterns during banana fruit ripening, with both genes significantly upregulated in tandem with ethylene production (Fig. 1). Critically, MaNAC19 also directly binds to MaACO1/13 promoters, and interacts with MaNAC029 to synergistically amplify transcriptional activation of MaACO1/13 (Figs. 2, 4, 5). These data demonstrate that MaNAC029 and MaNAC19 constitute a transcriptional regulatory module that fine-tunes ethylene biosynthesis through feedback regulation during banana fruit ripening, thereby expanding the transcriptional network governing ethylene-mediated ripening.
Ethylene-induced banana ripening orchestrates a cascade of physiological and biochemical alterations, including starch degradation, soluble sugar accumulation, aroma volatile biosynthesis, and chlorophyll catabolism, which collectively determine the commercial quality attributes of the fruit[26,27]. Sweetness intensity, chiefly determined by soluble sugars such as sucrose, serves as the primary determinant of consumer preference for banana fruit during ripening[33,34]. Previous studies have identified SPS as a pivotal enzyme catalyzing starch-to-sucrose conversion during banana fruit ripening[35], yet its upstream regulatory machinery remains poorly characterized. NAC TFs play pivotal roles in regulating fruit ripening and quality attributes[15,36]. Genome-wide identification and expression profiling revealed that NAC TFs exhibit distinct spatiotemporal expression dynamics during ethylene-induced banana fruit ripening, indicating their potential roles as transcriptional hubs in this regulatory process[36]. We previously found that MaNAC19 directly transcriptionally activates MaSPS1, thereby promoting sucrose synthesis throughout banana ripening[30]. This study reveals that MaNAC029, an upstream transcriptional modulator of ethylene biosynthesis[29], physically interacts with MaNAC19 to cooperatively govern the expression of ethylene synthesis genes and the sucrose synthase gene MaSPS1 (Figs. 2, 3, 5). Transcriptional analysis revealed that the expression of MaNAC029 and MaNAC19 peaked concurrently with the ethylene burst during early ripening and remained high thereafter (Fig. 1), facilitating sustained sucrose metabolism and accumulation throughout the ripening process, indicating their coordinated action is a key mechanism in ethylene-mediated sucrose synthesis. These findings establish the MaNAC029-MaNAC19 module as a central regulatory hub orchestrating ethylene and sucrose synthesis during banana fruit ripening. It is noteworthy that, although our results from the tobacco DLR transient expression system show a stronger activation of the MaSPS1 promoter upon co-expression of MaNAC029 and MaNAC19 compared to single expression (Fig. 5b), it remains essential for future work to confirm this synergistic regulation of sucrose accumulation in banana fruit.
The ripening of banana fruit induced by ethylene during the postharvest phase can be disrupted by adverse environmental conditions[37], such as extreme temperatures, which impair the accumulation of sugars and other flavor compounds[38]. MaNAC029 and MaNAC19 function both independently and synergistically to feedback-regulate ethylene and sucrose synthesis (Figs. 3−5). This dual capability forms a multifaceted, parallel regulatory system, which likely enhances the fruit's ethylene response efficiency and ensures the stable progression of ethylene-mediated quality development, thereby safeguarding it against fluctuations in the external environment. A fundamental challenge for future research is to elucidate the precise mechanisms through which the MaNAC029-MaNAC19 module orchestrates the dynamic regulation of ripening and quality under diverse environmental stresses.
Beyond PPIs, TFs can form transcriptional cascade modules to hierarchically regulate gene expression networks. For instance, MaSPL16 orchestrates banana fruit ripening by directly activating MaNAC029, forming a transcriptional cascade that coordinates ethylene biosynthesis[28]. Therefore, future investigations should delineate the upstream-downstream regulatory relationships of MaNAC029/19 and determine whether MaSPL16 functions as a common upstream regulator for both TFs.
-
Based on integrated evidence from this study and prior findings, a mechanistic model delineating how the MaNAC029-MaNAC19 heterodimeric transcriptional module orchestrates banana fruit ripening is proposed (Fig. 6). During the ethylene-induced postharvest ripening of banana fruit, the expression of MaNAC029 and MaNAC19 is activated first. MaNAC029 physically interacts with MaNAC19 to assemble a functional heterodimeric complex, which synergistically trans-activates ethylene biosynthesis genes MaACO1/13, thereby amplifying feedback-driven ethylene synthesis. Concurrently, this module cooperatively induces the sucrose phosphate synthase gene MaSPS1, enhancing sucrose accumulation to regulate banana fruit quality formation. Collectively, our findings establish the MaSPL16-MaNAC029 transcriptional hub as a central node integrating ethylene biosynthesis with sucrose metabolism, providing novel insights into transcriptional coordination of fruit ripening and quality.
This study was funded by the National Natural Science Foundation of China (32322075 and 32072279), and the China Agriculture Research System of MOF and MARA (CARS-31).
-
The authors confirm their contributions to the paper as follows: research design, did most of the experiments, and manuscript writing and revision: Shan W, Zhang YM; data analysis: Zhang YM, Wei W, Kuang JF, Chen JY, Lu WJ, Shan W. All authors reviewed the results and approved the final version of the manuscript.
-
Generated/analyzed data are included in this published article and its supplementary materials.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 Primers used in the study.
- Copyright: © 2026 by the author(s). Published by Maximum Academic Press on behalf of Chongqing University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang YM, Wei W, Kuang JF, Lu WJ, Chen JY, et al. 2026. MaNAC029 and MaNAC19 synergistically regulate ethylene and sucrose synthesis during banana fruit ripening. Plant Hormones 2: e002 doi: 10.48130/ph-0025-0029
MaNAC029 and MaNAC19 synergistically regulate ethylene and sucrose synthesis during banana fruit ripening
- Received: 02 November 2025
- Revised: 10 December 2025
- Accepted: 23 December 2025
- Published online: 27 January 2026
Abstract: Ethylene is a critical phytohormone for climacteric fruits, such as banana, triggering the initiation of ripening and subsequent physiological and metabolic shifts, ultimately facilitating the formation of characteristic fruit quality attributes. NAC transcription factors (TFs) are crucial in ethylene-induced fruit ipening and quality formation. Our recent study revealed that MaNAC029 orchestrates ethylene biosynthesis during banana fruit ripening by directly transcriptionally modulating ethylene synthesis genes (MaACO1/13). Meanwhile, MaNAC19 regulates sucrose accumulation via activating the sucrose phosphate synthase gene (MaSPS1), but the functional interplay between MaNAC029 and MaNAC19 has not been elucidated. Herein, MaNAC029/19 were found to exhibit synchronized expression patterns during banana fruit ripening, with both genes significantly upregulated following ethylene production. Protein-protein interaction analysis manifested that MaNAC029 physically interacts with MaNAC19. Critically, MaNAC029 binds specifically to the MaSPS1 promoter, while MaNAC19 occupies promoters of MaACO1/13, indicating cross-regulation of each other's downstream targets. Notably, MaNAC029 and MaNAC19 synergistically activate the promoter activities of MaACO1/13 and MaSPS1. Collectively, these results establish the MaNAC029-MaNAC19 complex as a cooperative transcriptional module that integrates ethylene biosynthesis and sucrose metabolism, advancing our mechanistic understanding of the transcriptional networks governing banana fruit ripening and quality formation.
-
Key words:
- Banana fruit /
- Ripening /
- NAC /
- Protein-protein interaction /
- TF regulatory module